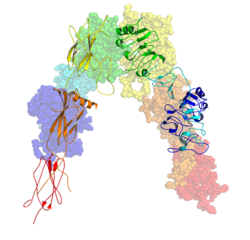
Insulin receptor
The insulin receptor (IR) is a transmembrane receptor that is activated by insulin, IGF-I, IGF-II and belongs to the large class of receptor tyrosine kinase.[5] Metabolically, the insulin receptor plays a key role in the regulation of glucose homeostasis; a functional process that under degenerate conditions may result in a range of clinical manifestations including diabetes and cancer.[6][7] Insulin signalling controls access to blood glucose in body cells. When insulin falls, especially in those with high insulin sensitivity, body cells begin only to have access to lipids that do not require transport across the membrane. So, in this way, insulin is the key regulator of fat metabolism as well. Biochemically, the insulin receptor is encoded by a single gene INSR, from which alternate splicing during transcription results in either IR-A or IR-B isoforms.[8] Downstream post-translational events of either isoform result in the formation of a proteolytically cleaved α and β subunit, which upon combination are ultimately capable of homo or hetero-dimerisation to produce the ≈320 kDa disulfide-linked transmembrane insulin receptor.[8]
Structure
[edit]Initially, transcription of alternative splice variants derived from the INSR gene are translated to form one of two monomeric isomers; IR-A in which exon 11 is excluded, and IR-B in which exon 11 is included. Inclusion of exon 11 results in the addition of 12 amino acids upstream of the intrinsic furin proteolytic cleavage site.

Upon receptor dimerisation, after proteolytic cleavage into the α- and β-chains, the additional 12 amino acids remain present at the C-terminus of the α-chain (designated αCT) where they are predicted to influence receptor–ligand interaction.[9]
Each isometric monomer is structurally organized into 8 distinct domains consists of; a leucine-rich repeat domain (L1, residues 1–157), a cysteine-rich region (CR, residues 158–310), an additional leucine rich repeat domain (L2, residues 311–470), three fibronectin type III domains; FnIII-1 (residues 471–595), FnIII-2 (residues 596–808) and FnIII-3 (residues 809–906). Additionally, an insert domain (ID, residues 638–756) resides within FnIII-2, containing the α/β furin cleavage site, from which proteolysis results in both IDα and IDβ domains. Within the β-chain, downstream of the FnIII-3 domain lies a transmembrane helix (TH) and intracellular juxtamembrane (JM) region, just upstream of the intracellular tyrosine kinase (TK) catalytic domain, responsible for subsequent intracellular signaling pathways.[10]
Upon cleavage of the monomer to its respective α- and β-chains, receptor hetero or homo-dimerisation is maintained covalently between chains by a single disulphide link and between monomers in the dimer by two disulphide links extending from each α-chain. The overall 3D ectodomain structure, possessing four ligand binding sites, resembles an inverted 'V', with the each monomer rotated approximately 2-fold about an axis running parallel to the inverted 'V' and L2 and FnIII-1 domains from each monomer forming the inverted 'V's apex.[10][11]
Ligand binding
[edit]

The insulin receptor's endogenous ligands include insulin, IGF-I and IGF-II. Using a cryo-EM, structural insight into conformational changes upon insulin binding was provided. Binding of ligand to the α-chains of the IR dimeric ectodomain shifts it from an inverted V-shape to a T-shaped conformation, and this change is propagated structurally to the transmembrane domains, which get closer, eventually leading to autophosphorylation of various tyrosine residues within the intracellular TK domain of the β-chain.[12] These changes facilitate the recruitment of specific adapter proteins such as the insulin receptor substrate proteins (IRS) in addition to SH2-B (Src Homology 2 - B ), APS and protein phosphatases, such as PTP1B, eventually promoting downstream processes involving blood glucose homeostasis.[14]
Strictly speaking the relationship between IR and ligand shows complex allosteric properties. This was indicated with the use of a Scatchard plots which identified that the measurement of the ratio of IR bound ligand to unbound ligand does not follow a linear relationship with respect to changes in the concentration of IR bound ligand, suggesting that the IR and its respective ligand share a relationship of cooperative binding.[15] Furthermore, the observation that the rate of IR-ligand dissociation is accelerated upon addition of unbound ligand implies that the nature of this cooperation is negative; said differently, that the initial binding of ligand to the IR inhibits further binding to its second active site - exhibition of allosteric inhibition.[15]
These models state that each IR monomer possesses 2 insulin binding sites; site 1, which binds to the 'classical' binding surface of insulin: consisting of L1 plus αCT domains and site 2, consisting of loops at the junction of FnIII-1 and FnIII-2 predicted to bind to the 'novel' hexamer face binding site of insulin.[5] As each monomer contributing to the IR ectodomain exhibits 3D 'mirrored' complementarity, N-terminal site 1 of one monomer ultimately faces C-terminal site 2 of the second monomer, where this is also true for each monomers mirrored complement (the opposite side of the ectodomain structure). Current literature distinguishes the complement binding sites by designating the second monomer's site 1 and site 2 nomenclature as either site 3 and site 4 or as site 1' and site 2' respectively.[5][14] As such, these models state that each IR may bind to an insulin molecule (which has two binding surfaces) via 4 locations, being site 1, 2, (3/1') or (4/2'). As each site 1 proximally faces site 2, upon insulin binding to a specific site, 'crosslinking' via ligand between monomers is predicted to occur (i.e. as [monomer 1 Site 1 - Insulin - monomer 2 Site (4/2')] or as [monomer 1 Site 2 - Insulin - monomer 2 site (3/1')]). In accordance with current mathematical modelling of IR-insulin kinetics, there are two important consequences to the events of insulin crosslinking; 1. that by the aforementioned observation of negative cooperation between IR and its ligand that subsequent binding of ligand to the IR is reduced and 2. that the physical action of crosslinking brings the ectodomain into such a conformation that is required for intracellular tyrosine phosphorylation events to ensue (i.e. these events serve as the requirements for receptor activation and eventual maintenance of blood glucose homeostasis).[14]
Applying cryo-EM and molecular dynamics simulations of receptor reconstituted in nanodiscs, the structure of the entire dimeric insulin receptor ectodomain with four insulin molecules bound was visualized, therefore confirming and directly showing biochemically predicted 4 binding locations.[13]
Agonists
[edit]A number of small-molecule insulin receptor agonists have been identified.[16]
Signal transduction pathway
[edit]The insulin receptor is a type of tyrosine kinase receptor, in which the binding of an agonistic ligand triggers autophosphorylation of the tyrosine residues, with each subunit phosphorylating its partner. The addition of the phosphate groups generates a binding site for the insulin receptor substrate (IRS-1), which is subsequently activated via phosphorylation. The activated IRS-1 initiates the signal transduction pathway and binds to phosphoinositide 3-kinase (PI3K), in turn causing its activation. This then catalyses the conversion of Phosphatidylinositol 4,5-bisphosphate into Phosphatidylinositol 3,4,5-trisphosphate (PIP3). PIP3 acts as a secondary messenger and induces the activation of phosphatidylinositol dependent protein kinase, which then activates several other kinases – most notably protein kinase B, (PKB, also known as Akt). PKB triggers the translocation of glucose transporter (GLUT4) containing vesicles to the cell membrane, via the activation of SNARE proteins, to facilitate the diffusion of glucose into the cell. PKB also phosphorylates and inhibits glycogen synthase kinase, which is an enzyme that inhibits glycogen synthase. Therefore, PKB acts to start the process of glycogenesis, which ultimately reduces blood-glucose concentration.[17]
-
Effect of insulin on glucose uptake and metabolism. Insulin binds to its receptor (1), which, in turn, starts many protein activation cascades (2). These include: translocation of Glut-4 transporter to the plasma membrane and influx of glucose (3), glycogen synthesis (4), glycolysis (5), and fatty acid synthesis (6).
-
Signal transduction of Insulin: At the end of the transduction process, the activated protein binds to the PIP2 phospholipids embedded in the membrane.
Function
[edit]Regulation of gene expression
[edit]The activated IRS-1 acts as a secondary messenger within the cell to stimulate the transcription of insulin-regulated genes. First, the protein Grb2 binds the P-Tyr residue of IRS-1 in its SH2 domain. Grb2 is then able to bind SOS, which in turn catalyzes the replacement of bound GDP with GTP on Ras, a G protein. This protein then begins a phosphorylation cascade, culminating in the activation of mitogen-activated protein kinase (MAPK), which enters the nucleus and phosphorylates various nuclear transcription factors (such as Elk1).
Stimulation of glycogen synthesis
[edit]Glycogen synthesis is also stimulated by the insulin receptor via IRS-1. In this case, it is the SH2 domain of PI-3 kinase (PI-3K) that binds the P-Tyr of IRS-1. Now activated, PI-3K can convert the membrane lipid phosphatidylinositol 4,5-bisphosphate (PIP2) to phosphatidylinositol 3,4,5-triphosphate (PIP3). This indirectly activates a protein kinase, PKB (Akt), via phosphorylation. PKB then phosphorylates several target proteins, including glycogen synthase kinase 3 (GSK-3). GSK-3 is responsible for phosphorylating (and thus deactivating) glycogen synthase. When GSK-3 is phosphorylated, it is deactivated, and prevented from deactivating glycogen synthase. In this roundabout manner, insulin increases glycogen synthesis.
Degradation of insulin
[edit]Once an insulin molecule has docked onto the receptor and effected its action, it may be released back into the extracellular environment or it may be degraded by the cell. Degradation normally involves endocytosis of the insulin-receptor complex followed by the action of insulin degrading enzyme. Most insulin molecules are degraded by liver cells. It has been estimated that a typical insulin molecule is finally degraded about 71 minutes after its initial release into circulation.[18]
Immune system
[edit]Besides the metabolic function, insulin receptors are also expressed on immune cells, such as macrophages, B cells, and T cells. On T cells, the expression of insulin receptors is undetectable during the resting state but up-regulated upon T-cell receptor (TCR) activation. Indeed, insulin has been shown when supplied exogenously to promote in vitro T cell proliferation in animal models. Insulin receptor signalling is important for maximizing the potential effect of T cells during acute infection and inflammation.[19][20]
Pathology
[edit]The main activity of activation of the insulin receptor is inducing glucose uptake. For this reason "insulin insensitivity", or a decrease in insulin receptor signaling, leads to diabetes mellitus type 2 – the cells are unable to take up glucose, and the result is hyperglycemia (an increase in circulating glucose), and all the sequelae that result from diabetes.
Patients with insulin resistance may display acanthosis nigricans.
A few patients with homozygous mutations in the INSR gene have been described, which causes Donohue syndrome or Leprechaunism. This autosomal recessive disorder results in a totally non-functional insulin receptor. These patients have low-set, often protuberant, ears, flared nostrils, thickened lips, and severe growth retardation. In most cases, the outlook for these patients is extremely poor, with death occurring within the first year of life. Other mutations of the same gene cause the less severe Rabson-Mendenhall syndrome, in which patients have characteristically abnormal teeth, hypertrophic gingiva (gums), and enlargement of the pineal gland. Both diseases present with fluctuations of the glucose level: After a meal the glucose is initially very high, and then falls rapidly to abnormally low levels.[21] Other genetic mutations to the insulin receptor gene can cause Severe Insulin Resistance.[22]
Interactions
[edit]Insulin receptor has been shown to interact with
References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000171105 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000005534 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b c Ward CW, Lawrence MC (April 2009). "Ligand-induced activation of the insulin receptor: a multi-step process involving structural changes in both the ligand and the receptor". BioEssays. 31 (4): 422–34. doi:10.1002/bies.200800210. PMID 19274663. S2CID 27645596.
- ^ Ebina Y, Ellis L, Jarnagin K, Edery M, Graf L, Clauser E, Ou JH, Masiarz F, Kan YW, Goldfine ID (April 1985). "The human insulin receptor cDNA: the structural basis for hormone-activated transmembrane signalling". Cell. 40 (4): 747–58. doi:10.1016/0092-8674(85)90334-4. PMID 2859121. S2CID 23230348.
- ^ Malaguarnera R, Sacco A, Voci C, Pandini G, Vigneri R, Belfiore A (May 2012). "Proinsulin binds with high affinity the insulin receptor isoform A and predominantly activates the mitogenic pathway". Endocrinology. 153 (5): 2152–63. doi:10.1210/en.2011-1843. PMID 22355074.
- ^ a b Belfiore A, Frasca F, Pandini G, Sciacca L, Vigneri R (October 2009). "Insulin receptor isoforms and insulin receptor/insulin-like growth factor receptor hybrids in physiology and disease". Endocrine Reviews. 30 (6): 586–623. doi:10.1210/er.2008-0047. PMID 19752219.
- ^ Knudsen L, De Meyts P, Kiselyov VV (December 2011). "Insight into the molecular basis for the kinetic differences between the two insulin receptor isoforms" (PDF). The Biochemical Journal. 440 (3): 397–403. doi:10.1042/BJ20110550. PMID 21838706.
- ^ a b Smith BJ, Huang K, Kong G, Chan SJ, Nakagawa S, Menting JG, Hu SQ, Whittaker J, Steiner DF, Katsoyannis PG, Ward CW, Weiss MA, Lawrence MC (April 2010). "Structural resolution of a tandem hormone-binding element in the insulin receptor and its implications for design of peptide agonists". Proceedings of the National Academy of Sciences of the United States of America. 107 (15): 6771–6. Bibcode:2010PNAS..107.6771S. doi:10.1073/pnas.1001813107. PMC 2872410. PMID 20348418.
- ^ McKern NM, Lawrence MC, Streltsov VA, Lou MZ, Adams TE, Lovrecz GO, Elleman TC, Richards KM, Bentley JD, Pilling PA, Hoyne PA, Cartledge KA, Pham TM, Lewis JL, Sankovich SE, Stoichevska V, Da Silva E, Robinson CP, Frenkel MJ, Sparrow LG, Fernley RT, Epa VC, Ward CW (September 2006). "Structure of the insulin receptor ectodomain reveals a folded-over conformation". Nature. 443 (7108): 218–21. Bibcode:2006Natur.443..218M. doi:10.1038/nature05106. PMID 16957736. S2CID 4381431.
- ^ a b Gutmann T, Kim KH, Grzybek M, Walz T, Coskun Ü (May 2018). "Visualization of ligand-induced transmembrane signaling in the full-length human insulin receptor". The Journal of Cell Biology. 217 (5): 1643–1649. doi:10.1083/jcb.201711047. PMC 5940312. PMID 29453311.
- ^ a b Gutmann T, Schäfer IB, Poojari C, Brankatschk B, Vattulainen I, Strauss M, Coskun Ü (January 2020). "Cryo-EM structure of the complete and ligand-saturated insulin receptor ectodomain". The Journal of Cell Biology. 219 (1). doi:10.1083/jcb.201907210. PMC 7039211. PMID 31727777.
- ^ a b c Kiselyov VV, Versteyhe S, Gauguin L, De Meyts P (February 2009). "Harmonic oscillator model of the insulin and IGF1 receptors' allosteric binding and activation". Molecular Systems Biology. 5 (5): 243. doi:10.1038/msb.2008.78. PMC 2657531. PMID 19225456.
- ^ a b de Meyts P, Roth J, Neville DM, Gavin JR, Lesniak MA (November 1973). "Insulin interactions with its receptors: experimental evidence for negative cooperativity". Biochemical and Biophysical Research Communications. 55 (1): 154–61. doi:10.1016/S0006-291X(73)80072-5. PMID 4361269.
- ^ Kumar L, Vizgaudis W, Klein-Seetharaman J (July 2022). "Structure-based survey of ligand binding in the human insulin receptor". Br J Pharmacol. 179 (14): 3512–3528. doi:10.1111/bph.15777. PMID 34907529. S2CID 245242018.
- ^ Berg JM, Tymoczko JL, Gatto GJ, Stryer L (2019). Biochemistry (9th ed.). W H Freeman/Macmillan International. p. 897. ISBN 978-1-319-11465-7.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Duckworth WC, Bennett RG, Hamel FG (October 1998). "Insulin degradation: progress and potential". Endocrine Reviews. 19 (5): 608–24. doi:10.1210/edrv.19.5.0349. PMID 9793760.
- ^ Tsai S, Clemente-Casares X, Zhou AC, Lei H, Ahn JJ, Chan YT, et al. (August 2018). "Insulin Receptor-Mediated Stimulation Boosts T Cell Immunity during Inflammation and Infection". Cell Metabolism. 28 (6): 922–934.e4. doi:10.1016/j.cmet.2018.08.003. PMID 30174303.
- ^ Fischer HJ, Sie C, Schumann E, Witte AK, Dressel R, van den Brandt J, Reichardt HM (March 2017). "The Insulin Receptor Plays a Critical Role in T Cell Function and Adaptive Immunity". Journal of Immunology. 198 (5): 1910–1920. doi:10.4049/jimmunol.1601011. PMID 28115529.
- ^ Longo N, Wang Y, Smith SA, Langley SD, DiMeglio LA, Giannella-Neto D (June 2002). "Genotype-phenotype correlation in inherited severe insulin resistance". Human Molecular Genetics. 11 (12): 1465–75. doi:10.1093/hmg/11.12.1465. PMID 12023989. S2CID 15924838.
- ^ Melvin A, Stears A (2017). "Severe insulin resistance: pathologies". Practical Diabetes. 34 (6): 189–194a. doi:10.1002/pdi.2116. S2CID 90238599. Retrieved 31 October 2020.
- ^ Maddux BA, Goldfine ID (January 2000). "Membrane glycoprotein PC-1 inhibition of insulin receptor function occurs via direct interaction with the receptor alpha-subunit". Diabetes. 49 (1): 13–9. doi:10.2337/diabetes.49.1.13. PMID 10615944.
- ^ Langlais P, Dong LQ, Hu D, Liu F (June 2000). "Identification of Grb10 as a direct substrate for members of the Src tyrosine kinase family". Oncogene. 19 (25): 2895–903. doi:10.1038/sj.onc.1203616. PMID 10871840. S2CID 25923169.
- ^ Hansen H, Svensson U, Zhu J, Laviola L, Giorgino F, Wolf G, Smith RJ, Riedel H (April 1996). "Interaction between the Grb10 SH2 domain and the insulin receptor carboxyl terminus". The Journal of Biological Chemistry. 271 (15): 8882–6. doi:10.1074/jbc.271.15.8882. PMID 8621530.
- ^ Liu F, Roth RA (October 1995). "Grb-IR: a SH2-domain-containing protein that binds to the insulin receptor and inhibits its function". Proceedings of the National Academy of Sciences of the United States of America. 92 (22): 10287–91. Bibcode:1995PNAS...9210287L. doi:10.1073/pnas.92.22.10287. PMC 40781. PMID 7479769.
- ^ He W, Rose DW, Olefsky JM, Gustafson TA (March 1998). "Grb10 interacts differentially with the insulin receptor, insulin-like growth factor I receptor, and epidermal growth factor receptor via the Grb10 Src homology 2 (SH2) domain and a second novel domain located between the pleckstrin homology and SH2 domains". The Journal of Biological Chemistry. 273 (12): 6860–7. doi:10.1074/jbc.273.12.6860. PMID 9506989.
- ^ Frantz JD, Giorgetti-Peraldi S, Ottinger EA, Shoelson SE (January 1997). "Human GRB-IRbeta/GRB10. Splice variants of an insulin and growth factor receptor-binding protein with PH and SH2 domains". The Journal of Biological Chemistry. 272 (5): 2659–67. doi:10.1074/jbc.272.5.2659. PMID 9006901.
- ^ Kasus-Jacobi A, Béréziat V, Perdereau D, Girard J, Burnol AF (April 2000). "Evidence for an interaction between the insulin receptor and Grb7. A role for two of its binding domains, PIR and SH2". Oncogene. 19 (16): 2052–9. doi:10.1038/sj.onc.1203469. PMID 10803466. S2CID 10955124.
- ^ Aguirre V, Werner ED, Giraud J, Lee YH, Shoelson SE, White MF (January 2002). "Phosphorylation of Ser307 in insulin receptor substrate-1 blocks interactions with the insulin receptor and inhibits insulin action". The Journal of Biological Chemistry. 277 (2): 1531–7. doi:10.1074/jbc.M101521200. PMID 11606564.
- ^ Sawka-Verhelle D, Tartare-Deckert S, White MF, Van Obberghen E (March 1996). "Insulin receptor substrate-2 binds to the insulin receptor through its phosphotyrosine-binding domain and through a newly identified domain comprising amino acids 591-786". The Journal of Biological Chemistry. 271 (11): 5980–3. doi:10.1074/jbc.271.11.5980. PMID 8626379.
- ^ O'Neill TJ, Zhu Y, Gustafson TA (April 1997). "Interaction of MAD2 with the carboxyl terminus of the insulin receptor but not with the IGFIR. Evidence for release from the insulin receptor after activation". The Journal of Biological Chemistry. 272 (15): 10035–40. doi:10.1074/jbc.272.15.10035. PMID 9092546.
- ^ Braiman L, Alt A, Kuroki T, Ohba M, Bak A, Tennenbaum T, Sampson SR (April 2001). "Insulin induces specific interaction between insulin receptor and protein kinase C delta in primary cultured skeletal muscle". Molecular Endocrinology. 15 (4): 565–74. doi:10.1210/mend.15.4.0612. PMID 11266508.
- ^ Rosenzweig T, Braiman L, Bak A, Alt A, Kuroki T, Sampson SR (June 2002). "Differential effects of tumor necrosis factor-alpha on protein kinase C isoforms alpha and delta mediate inhibition of insulin receptor signaling". Diabetes. 51 (6): 1921–30. doi:10.2337/diabetes.51.6.1921. PMID 12031982.
- ^ Maegawa H, Ugi S, Adachi M, Hinoda Y, Kikkawa R, Yachi A, Shigeta Y, Kashiwagi A (March 1994). "Insulin receptor kinase phosphorylates protein tyrosine phosphatase containing Src homology 2 regions and modulates its PTPase activity in vitro". Biochemical and Biophysical Research Communications. 199 (2): 780–5. doi:10.1006/bbrc.1994.1297. PMID 8135823.
- ^ Kharitonenkov A, Schnekenburger J, Chen Z, Knyazev P, Ali S, Zwick E, White M, Ullrich A (December 1995). "Adapter function of protein-tyrosine phosphatase 1D in insulin receptor/insulin receptor substrate-1 interaction". The Journal of Biological Chemistry. 270 (49): 29189–93. doi:10.1074/jbc.270.49.29189. PMID 7493946.
- ^ Kotani K, Wilden P, Pillay TS (October 1998). "SH2-Balpha is an insulin-receptor adapter protein and substrate that interacts with the activation loop of the insulin-receptor kinase". The Biochemical Journal. 335 (1): 103–9. doi:10.1042/bj3350103. PMC 1219757. PMID 9742218.
- ^ Nelms K, O'Neill TJ, Li S, Hubbard SR, Gustafson TA, Paul WE (December 1999). "Alternative splicing, gene localization, and binding of SH2-B to the insulin receptor kinase domain". Mammalian Genome. 10 (12): 1160–7. doi:10.1007/s003359901183. PMID 10594240. S2CID 21060861.
Further reading
[edit]- Pearson RB, Kemp BE (1991). "[3] Protein kinase phosphorylation site sequences and consensus specificity motifs: Tabulations". Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods in Enzymology. Vol. 200. pp. 62–81. doi:10.1016/0076-6879(91)00127-I. ISBN 9780121821012. PMID 1956339.
- Joost HG (February 1995). "Structural and functional heterogeneity of insulin receptors". Cellular Signalling. 7 (2): 85–91. doi:10.1016/0898-6568(94)00071-I. PMID 7794689.
- O'Dell SD, Day IN (July 1998). "Insulin-like growth factor II (IGF-II)". The International Journal of Biochemistry & Cell Biology. 30 (7): 767–71. doi:10.1016/S1357-2725(98)00048-X. PMID 9722981.
- Lopaczynski W (1999). "Differential regulation of signaling pathways for insulin and insulin-like growth factor I". Acta Biochimica Polonica. 46 (1): 51–60. doi:10.18388/abp.1999_4183. PMID 10453981.
- Sasaoka T, Kobayashi M (August 2000). "The functional significance of Shc in insulin signaling as a substrate of the insulin receptor". Endocrine Journal. 47 (4): 373–81. doi:10.1507/endocrj.47.373. PMID 11075717.
- Perz M, Torlińska T (2001). "Insulin receptor--structural and functional characteristics". Medical Science Monitor. 7 (1): 169–77. PMID 11208515.
- Benaim G, Villalobo A (August 2002). "Phosphorylation of calmodulin. Functional implications". European Journal of Biochemistry. 269 (15): 3619–31. doi:10.1046/j.1432-1033.2002.03038.x. hdl:10261/79981. PMID 12153558.
External links
[edit]- Insulin+receptor at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Overview of all the structural information available in the PDB for UniProt: P06213 (Insulin receptor) at the PDBe-KB.