User:Biochem153aj/sandbox
First and Other Edits
[edit]Fixed a sentence in "Tyrosine"
- 23:49, 28 April 2019 diff hist -1 m Tyrosine Joined two sentences together into one. Tag: Visual edit
- 23:49, 28 April 2019 Biochem153aj talk contribs m 23,834 bytes -1 Joined two sentences together into one. undo Tag: Visual edit
Fixed a sentence in the leading paragraph for "Glycolysis."
- curprev 05:25, 21 May 2019 Biochem153aj talk contribs m 73,324 bytes -2 undo Tag: Visual edit
- curprev 05:25, 21 May 2019 Biochem153aj talk contribs m 73,326 bytes +61 undo Tag: Visual edit
Peer Reviewed Articles
[edit]Kataegis
[edit]Hi Aureliall, I will be reviewing your article. Here are my edits listed below.
1. Does the article flow well? Well organized? The article flows relatively well. There are several things that I would suggest. I would shorten the introduction and move part of it to a completely new section. I would group the sentences where you describe the "Greek origin" of the word into an entirely new section as well. This has no relation to why the reader would care to look up kataegis. This is a rather superfluous detail to include in the main introduction. Another suggestion would be to change the tense of the entire passage to be the same, particularly present, tense. There are both present and past tense verbs, making it particularly difficult to read. For organization, please see the suggestions above on grouping. Also, you do not have to generate your own reference section. This is automatically performed by Wikipedia when you add a citation.
2. Is the level of detail appropriate? Not too much or too little? The level of detail seems appropriate. However, I would cut down on the origins of the word, and move it to a different section. I would like to see more detail for clinical significance, perhaps.
3. Well organized: is content in the appropriate section and not redundant? Yes. This is a strong point in your article. I found little to no redundancies in your sections. Each offered new information.
4. Does each section stand alone? Yes. But I would try to connect them to the overall section more. For example, try to explain more why APOBEC enzyme family and TLS DNA polymerase are considered "mechanisms."
5. Is it neutral? Yes. I did not detect any bias.
6. Is everything cited? Yes. But don't forget to fix the reference section, as aforementioned.
7. Are there grammatical errors? Yes. There are quite a few. Several have to do with singular and plural usage. Others have to do with matching tenses. For example "Compare to other cancer-related mutations, such..." should be "Compared..."
8. What images would be useful? Some images regarding the APOBEC enzyme family perhaps, or a graphic showing the changes from either C→T or G→T. The protein data base should also have an image for TLS DNA polymerase and Rev1.
9. All images are explained clearly? Yes. The rainfall plot is explained quite well. But please delete the "we."
10. Is it clear? Yes and no. Although the level of detail is appropriate, it could be explained more clearly. The organization does need some work.
11. Is there irrelevant information, or relevant info missing? Perhaps the origins of the word could be deemed irrelevant but this is a personal choice. However, clinical significance does seem understudied and does need more information.
12. Scientific inaccuracy Whatever was presented seemed accurate and there were no glaring errors that I found.
Overall, this was a very good article. Some pictures, fixing of grammatical errors, and better organization would help to improve this. Hope my suggestions help! Biochem153aj (talk) 05:44, 22 May 2019 (UTC)
Steady State (Biochemistry)
[edit]1.Does the article flow well? Well Organized? Yes. The article is organized well into different sections. However, the sections are not clearly defined but are named rather generically (i.e. Blood Glucose, or Nitrogen-Containing Molecules). They do not specify the importance or relation of that topic to the broader concept of “Steady State.” For flow, citations are not needed after every single sentence, but actually after the passage of which the reference is being used. All the citations after each sentence actually hinders the reader. For example, [1] in the paragraphs under “Article Rough Draft” should be after each paragraph, not after each sentence. Steady state is explained well, as the article moves from overall description to a more specific definition of energy carriers which then play an important role in different pathways.
2.Is the level of detail appropriate? Not too much or too little? The level of detail is somewhat appropriate. However, the way that it is written is not with the appropriate scientific jargon. The detail is generalized in several areas and can lead to oversimplification of topics. For example, the description of blood lactate levels was lacking in certain areas and molecules.
3.Well organized: is content in the appropriate section and not redundant? As aforementioned, the content was well organized. But there was some repetition in wording. For example, for the Blood glucose and Nitrogen Containing Molecule sections, for each paragraph, the beginning sentences were very similar.
4.Does each section stand alone? Yes. The sections do not overlap in information. However, more detail should be included.
5.Is it neutral? Somewhat. Several times throughout the article, a statement will be made and then a simple example will be given in parentheses. However, this example seems irrelevant and simplified compared to the topic being subscribed. This problem may be alleviated by changing the writing to fit a more scientific tone, perhaps.
6.Is everything cited? Yes. However, please fix format.
7.Are there grammatical errors? No. But structure of sentences could be fixed. For example, this sentence: “In one step of the glycolysis pathway catalyzed by PFK-1, the equilibrium constant of reaction is about 1000…” This should be worded different. “About 1000” is incorrect. It should be “approximately 1000.” This is just a small example of a larger problem.
8.What images would be useful? Images of the energy pathways. Examples of steady state systems with graphics. For example, in lecture, we had a graphic at how blood glucose is maintained in the body at steady levels. Something similar to this would be good.
9.All images are explained clearly No images present currently.
10.Is it clear? Yes. Understanding the information present was very easy. This article is clear to understand.
11.Is there irrelevant information, or relevant info missing? No. All the information is related to what a “steady-state system” is.
12.Scientific inaccuracy No scientific inaccuracy. Just sentence structure could be improved.
Overall, very good article. Easy to read, clear, cited, and good explanations. Just a few suggestions: fix some of the wording and the citations and add images and you should be good to go! Hope these suggestions help!. Biochem153aj (talk) 07:59, 22 May 2019 (UTC)
Week 5 Reflection: Did not know to post on CCLE
[edit]Proteins/Lipids/Carbohydrates
Week 5 Reflection: Article does not have much information. Even other journals that I find have very little information on Propionyl-CoA but mostly focus on the carboxylase.
New Reflection After seeing the prompt: Write about your introduction to wikipedia. What did you expect? How has the experience met your expectations and how is it different? Include something you thought was really interesting about editing wikipedia or that you have learned during this process.
Wikipedia is so vast, it is unbelievable. I did not except so much collaboration. The experience has far exceeded my expectations as so many people are working together to evaluate the veracity of information. I thought what was interesting that editors are very respectful of one another, even with conflicting information. This leads to my second point: any inaccuracies of information should not be a problem for "outside" readers. There are so many people working for the betterment of wikipedia, any small incorrect detail will somehow be discovered one way or another. As I previously stated, I did not except wikipedia to be so popular with contributions. It seems more people contribute than actually read the articles for information!
Miscellaneous
[edit]Women Biochemists
Molecular Cell Biology
Wikiproject
Category:biochemistry
product inhibition
chemiosmotic theory
enzymes in glycolysis, TCA cycle, oxidative phosphorylation. some need room for expansion
amino acid articles. taste receptors.
Proteins
[edit]Revised Outline
[edit]Outline for addition
- Add reaction with Propionyl-CoA carboxylase
- include picture and short description
- Metabolic intermediates
- Expand on production through beta-oxidation of odd chain fatty acids
- add reaction
- include figures
- link: https://www.ncbi.nlm.nih.gov/books/NBK22387/
- add reaction
- expand on production by metabolism of isoleucine and valine
- Expand on different mechanisms
- Production from conversion of cholesterol to bile acids
- Expand on production from conversion of cholesterol to bile acids
- Metabolism in Bacteria
New Section for Draft
- Methylcitrate Cycle
- Overview
- Myobacterium Tuberculosis Metabolism
- include picture of reaction with intermediates
New Section-Expansion on Plant Metabolism
- In Arabidopsis
Human Uses Other than TCA Cycle
- Human Gene-Gen5
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4932917/
- include picture from pymol
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4932917/
Lead Paragraph
[edit]Propionyl-CoA is a coenzyme A derivative of propionic acid. It is composed of a 24 total carbon chain (without the coenzyme, it is a 3 carbon structure) and its production and metabolic fate depend on which organism it is present in.[1] Several different pathways can lead to its production, such as through the catabolism of specific amino acids or the oxidation of odd-chain fatty acids.[2] It later can be broken down by propionly-CoA carboxylase or through the methylcitrate cycle.[3] In different organisms, however, propionyl-CoA can be sequestered into controlled regions, to alleviate its potential toxicity through accumulation.[4] Genetic deficiencies regarding the production and breakdown of propionyl-CoA also have great human significance.[5]
Propionyl-CoA Production
[edit]There are several different pathways through which propionyl-CoA can be be produced:
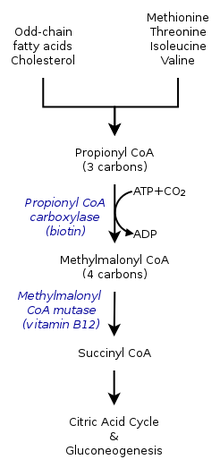
- Propionyl-CoA, a three-carbon structure, is considered to be a minor species. Therefore, odd-number chains of fatty acids are oxidized to yield both propionyl-CoA as well as acetyl-CoA. Propionyl-CoA is later converted into succinyl-CoA through propionyl-CoA carboxylase (PCC) by the use of vitamin B12.[2]
- Propionyl-CoA is not only produced from the oxidation of odd-chain fatty acids, but also by the catabolism of amino acids: methionine, valine, isoleucine, and threonine. Furthermore, catabolism of amino acids can also be a result of the conversion of propionyl-CoA to methylmalonyl-CoA by propionyl-CoA carboxylase.[1]
- Cholesterol oxidation, which forms bile acids, also forms propionyl-CoA as a side product. In an experiment performed by Suld et al., when combining liver mitochondria and propionic acid with the addition of coenzyme A, labeled isotopes of psionic acid were degraded. However, following 5β-cholestane-3α,7α,12α,26-tetrol-26,27-C14 incubation, propionyl CoA was able to be rescued along with the formation of bile.[6]
Metabolic Fate
[edit]The metabolic (catabolic fate) of propionyl-CoA depends on what environment it is being synthesized in. Therefore, propionyl-CoA in an anaerobic environment could have a different fate than in an aerobic organism. The multiple pathways, either catabolism by propionyl-CoA carboxylase or methylcitrate synthase also depend on the presence of the genes.[7]
Reaction with Propionyl-CoA carboxylase:
[edit]
Within the citric acid cycle, propionyl-CoA, which interacts with methylcitrate, is also catalyzed into methylmalonyl-CoA through carboxylation by propionyl-CoA carboxylase (PCC). Methylmalonyl-CoA is later transformed to succinyl-CoA to be further used in the tricarboxylic acid cycle. PCC not only catalyzes the carboxylation of propionyl-CoA to methylmalonyl-CoA, but also acts on several different acyl-CoAs. Nevertheless, it's highest binding affinity is to propionyl-CoA. It was further shown that propionyl-CoA transformation is inhibited by the absence of several TCA markers, such as glutamate. The mechanism is shown by the figure to the left.[2]
Methylcitrate Cycle
[edit]Propionyl-CoA accumulation can prove toxic to different organisms. Since different cycles have been proposed regarding how propionyl-CoA is transformed into pyruvate, one studied mechanism is the methylcitrate cycle. The initial reaction is beta-oxidation to form the propionyl-CoA which is further broken down by the cycle. This pathway involves the enzymes methylcitrate synthase, methylcitrate dehyratase, methylisocitrate lyase, succinate dehydrogenase, fumarase, citrate synthase, aconitase, malate:quinone oxidoreductase, isocitrate lyase, and malate synthase. These all contribute to the overall reaction to detoxify the bacteria from harmful propionyl-CoA. It is also attributed as a resulting pathway due to the catabolism of fatty acids in mycobacteria.[3] In order to proceed, the prpC gene codes for methylcitrate synthase, and if not present, the methylcitrate cycle will not occur. Instead, catabolism proceeds through propionyl-CoA carboxylase.[7]
Bacterial Metabolism
[edit]
Myobacterium Tuberculosis Metabolism
[edit]The oxidation of propionyl-CoA to form pyruvate is influenced by its necessity in mycobacterium tuberculosis. Accumulation of propionyl-CoA can lead to toxic effects. In mycobacterium tuberculosis, it has been suggested that the metabolism of propionyl-CoA is involved in cell wall biogenesis. A lack of such catabolism would therefore increase the susceptibility of the cell to various toxins, particularly to macrophage antimicrobial mechanisms. Another hypothesis regarding the fate of propionyl-CoA in M. tuberculosis is that since propionyl-CoA is produced by beta odd chain fatty acid catabolism, the methylcitrate cycle is activated subsequently to negate any potential toxicity, acting as a buffering mechanism.[10]
Possible Sequestration in R. Sphaeroides
[edit]Propionyl-CoA has can have many adverse and toxic affects on different species, including bacterium. For example, inhibition of pyruvate dehydrogenase by an accumulation of propionyl-CoA in Rhodobacter sphaeroides and can prove deadly. Furthermore, as with E. coli, an influx of propionyl-CoA in Myobacterial species. can result in toxicity if not dealt with immediately. This toxicity is caused by a pathway involving the lipids that form the bacterial cell wall. Using esterification of long-chain fatty acids, excess propionyl-CoA can be sequestered and stored in the lipid, triacylglycerol (TAG), leading to regulation of elevated propionyl-CoA levels.[4]
Escherichia Coli Metabolism
[edit]In an investigation performed by Luo et al., Escherichia coli strains were utilized to examine how the metabolism of propionyl-CoA could potentially lead to the production of 3-hydroxypropionic acid (3-HP). It was shown that a mutation in a key gene involved in the pathway, succinate CoA-transferase, led to a significant increase in 3-HP. However, this is still a developing field and information on this topic is limited.[7][11]

Plant Metabolism
[edit]Amino acid metabolism in plants has been deemed a controversial topic, due to the lack of concrete evidence for any particular pathway. However, it has been suggested that enzymes related to the production and use of propionyl-CoA are involved. Associated with this is the metabolism of isobutyrl-CoA. These two molecules are deemed to be intermediates in valine metabolism. As propionate consists in the form of propionyl-CoA, it was discovered that propionyl-CoA is converted to β-hydroxypropionate through a peroxisomal enzymatic β-oxidation pathway. Nevertheless, in the plant Arabidopsis, key enzymes in the conversion of valine to propionyl-CoA were not observed. Through different experiments performed by Lucas et al., it has been suggested that in plants, through peroxisomal enzymes, propionyl-CoA (and isobutyrl-CoA) are involved in the metabolism of many different substrates (currently being evaluated for identity), and not just valine.[12]
Fungi Metabolism
[edit]
Propionyl-CoA production through the catabolism of fatty acids is also associated with thioesterifcation. In a study concerning Aspergillus nidulans, it was found that with the inhibition of a methylcitrate synthase gene, mcsA, of the pathway described above, production of distinct polyketides was inhibited as well. Therefore, the utilization of propionyl-CoA through the methylcitrate cycle decreases its concentration, while subsequently increasing the concentration of polyketides. A polyketide differs from the more commonly known compound, the polypeptide, in that a polyketide is a structure commonly found in fungi that is made of acetyl- and malonyl-CoAs. They hold certain properties that have increased the research into their medicinal and potentially harmful attributes, by limiting polyketide toxicity to crops in agriculture through phytopathogenic fungi.[13]
Human and Clinical Significance
[edit]Gen5
[edit]
Similar to how plant peroxisomal enzymes bind propionyl-CoA and isobutyrl-CoA, Gen5, an acetyltransferase in humans, binds to propionyl-CoA and butyl-CoA. These specifically bind to the catalytic domain of Gen5L2. This conserved acetyltransferase is responsible for the regulation of transcription by lysine acetylation of the histone N-terminal tails. This function of acetylation has a much higher reaction rate than propionylation or butyrylation. Because of the structure of propionyl-CoA, Gen5 distinguishes between different acyl-CoA molecules. In fact, it was found that the propyl group of butyrl-CoA cannot bind due to lack of stereospecificity to the active binding site of Gen5 due to the unsaturated acyl chains. On the other hand, the C3 of propionyl-CoA can fit into the active site of Gen5 with the correct orientation.[14]
Propionic Acidemia
[edit]In the neonatal developmental stages, propionic acidemia, which is a medical issue defined as the lack of propionyl-CoA carboxylase, can cause impairment, mental disability, and numerous other issues. This is caused by an accumulation of propionyl-CoA because it cannot be converted to methylmalonyl-CoA. Newborns are tested for elevated propionylcarnitine. Further ways of diagnosing this disease could include urine samples. Medications used help to reverse and prevent further symptoms by including supplements to decrease propionate production.[15]
References
[edit]- ^ a b Dasgupta, Amitava (2019-01-01), Dasgupta, Amitava (ed.), "Chapter 2 - Biotin: Pharmacology, Pathophysiology, and Assessment of Biotin Status", Biotin and Other Interferences in Immunoassays, Elsevier, pp. 17–35, ISBN 9780128164297, retrieved 2019-06-03
- ^ a b c Wongkittichote, Parith; Ah Mew, Nicholas; Chapman, Kimberly A. (2017-12-01). "Propionyl-CoA carboxylase – A review". Molecular Genetics and Metabolism. 122 (4): 145–152. doi:10.1016/j.ymgme.2017.10.002. ISSN 1096-7192.
- ^ a b Upton, Anna M.; McKinney, John D. (2007). "Role of the methylcitrate cycle in propionate metabolism and detoxification in Mycobacterium smegmatis". Microbiology. 153 (12): 3973–3982. doi:10.1099/mic.0.2007/011726-0.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Dolan, Stephen K.; Wijaya, Andre; Geddis, Stephen M.; Spring, David R.; Silva-Rocha, Rafael; Welch, Martin (2018). "Loving the poison: the methylcitrate cycle and bacterial pathogenesis". Microbiology. 164 (3): 251–259. doi:10.1099/mic.0.000604.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Shchelochkov, Oleg A.; Carrillo, Nuria; Venditti, Charles (1993), Adam, Margaret P.; Ardinger, Holly H.; Pagon, Roberta A.; Wallace, Stephanie E. (eds.), "Propionic Acidemia", GeneReviews®, University of Washington, Seattle, PMID 22593918, retrieved 2019-06-03
- ^ Suld, Helga; Staple, Ezra; Gurin, Samuel (July 1961). "Mechanism of Formation of Bile Acids from Cholesterol: Oxidation of 5BCholestane-3a,7a,12a-trio1 and Formation of Propionic Acid from the Side Chain by Rat Liver Mitochondria" (PDF). The Journal of Biological Chemistry. 237: 338–344 – via JBC.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c Chang, Yanhong; Quiroga-Sánchez, Diego Leandro; Nie, Zhihua; Liu, Xiaohui; Zhou, Dafeng; Luo, Hui (2016-05-26). "Production of 3-Hydroxypropionic Acid via the Propionyl-CoA Pathway Using Recombinant Escherichia coli Strains". PLOS ONE. 11 (5): e0156286. doi:10.1371/journal.pone.0156286. ISSN 1932-6203. PMC 4882031. PMID 27227837.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: unflagged free DOI (link) - ^ Liu, Xin-Xin; Liu, Weibing; Shen, Meng-Jia; Ye, Bang-Ce (2018-06-21). "Nitrogen regulator GlnR directly controls transcription of prpDBC operon involved in methylcitrate cycle in Mycobacterium smegmatis". dx.doi.org. Retrieved 2019-06-13.
- ^ Ryan, Dylan G.; Murphy, Michael P.; Frezza, Christian; Prag, Hiran A.; Chouchani, Edward T.; O’Neill, Luke A.; Mills, Evanna L. (2018-12-30). "Coupling Krebs cycle metabolites to signalling in immunity and cancer". Nature Metabolism. 1 (1): 16–33. doi:10.1038/s42255-018-0014-7. ISSN 2522-5812.
- ^ Muñoz‐Elías, Ernesto J.; Upton, Anna M.; Cherian, Joseph; McKinney, John D. (2006). "Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulence". Molecular Microbiology. 60 (5): 1109–1122. doi:10.1111/j.1365-2958.2006.05155.x. ISSN 1365-2958.
- ^ Xiang, Hua; Zhou, Jian; Zhao, Dahe; Cai, Lei; Zhang, Siliang; Wang, Zejian; Wang, Lei; Liu, Hailong; Cai, Shuangfeng (2013-05-01). "Multiple Propionyl Coenzyme A-Supplying Pathways for Production of the Bioplastic Poly(3-Hydroxybutyrate-co-3-Hydroxyvalerate) in Haloferax mediterranei". Applied and Environmental Microbiology. 79 (9): 2922–2931. doi:10.1128/AEM.03915-12. ISSN 0099-2240. PMID 23435886.
- ^ Hawes, John W.; Graybill, Eric R.; Erb, Jeremy M.; Filley, Jessica R.; Lucas, Kerry A. (2007-08-24). "Peroxisomal Metabolism of Propionic Acid and Isobutyric Acid in Plants". Journal of Biological Chemistry. 282 (34): 24980–24989. doi:10.1074/jbc.M701028200. ISSN 0021-9258. PMID 17580301.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Keller, Nancy P.; Brock, Matthias; Zhang, Yong-Qiang (2004-10-01). "Connection of Propionyl-CoA Metabolism to Polyketide Biosynthesis in Aspergillus nidulans". Genetics. 168 (2): 785–794. doi:10.1534/genetics.104.027540. ISSN 0016-6731. PMID 15514053.
- ^ Wolberger, C.; Ringel, A. E. (2016-07-01). "Structural basis for acyl-group discrimination by human Gcn5L2". Acta Crystallographica Section D: Structural Biology. 72 (7): 841–848. doi:10.1107/S2059798316007907. ISSN 2059-7983.
- ^ Shchelochkov, Oleg A.; Carrillo, Nuria; Venditti, Charles (1993), Adam, Margaret P.; Ardinger, Holly H.; Pagon, Roberta A.; Wallace, Stephanie E. (eds.), "Propionic Acidemia", GeneReviews®, University of Washington, Seattle, PMID 22593918, retrieved 2019-06-03
