UDP-glucose 4-epimerase
| UDP-glucose 4-epimerase | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | UDPgalactose 4-epimerase4-epimeraseuridine diphosphate glucose 4-epimeraseUDPG-4-epimeraseUDP-galactose 4-epimeraseuridine diphosphoglucose epimeraseuridine diphospho-galactose-4-epimeraseUDP-D-galactose 4-epimeraseUDP-glucose epimeraseuridine diphosphoglucose 4-epimeraseuridine diphosphate galactose 4-epimerase | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | GeneCards: [1]; OMA:- orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| UDP-glucose 4-epimerase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
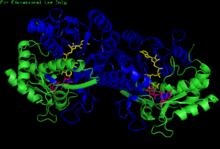 H. sapiens UDP-glucose 4-epimerase homodimer bound to NADH and UDP-glucose. Domains: N-terminal and C-terminal. | |||||||||
| Identifiers | |||||||||
| EC no. | 5.1.3.2 | ||||||||
| CAS no. | 9032-89-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| UDP-galactose-4-epimerase | |||||||
|---|---|---|---|---|---|---|---|
 Human GALE bound to NAD+ and UDP-GlcNAc, with N- and C-terminal domains highlighted. Asn 207 contorts to accommodate UDP-GlcNAc within the active site. | |||||||
| Identifiers | |||||||
| Symbol | GALE | ||||||
| NCBI gene | 2582 | ||||||
| HGNC | 4116 | ||||||
| OMIM | 606953 | ||||||
| RefSeq | NM_000403 | ||||||
| UniProt | Q14376 | ||||||
| Other data | |||||||
| EC number | 5.1.3.2 | ||||||
| Locus | Chr. 1 p36-p35 | ||||||
| |||||||
| NAD-dependent epimerase/dehydratase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | ? | ||||||||
| Pfam | PF01370 | ||||||||
| InterPro | IPR001509 | ||||||||
| Membranome | 330 | ||||||||
| |||||||||
The enzyme UDP-glucose 4-epimerase (EC 5.1.3.2), also known as UDP-galactose 4-epimerase or GALE, is a homodimeric epimerase found in bacterial, fungal, plant, and mammalian cells. This enzyme performs the final step in the Leloir pathway of galactose metabolism, catalyzing the reversible conversion of UDP-galactose to UDP-glucose.[1] GALE tightly binds nicotinamide adenine dinucleotide (NAD+), a co-factor required for catalytic activity.[2]
Additionally, human and some bacterial GALE isoforms reversibly catalyze the formation of UDP-N-acetylgalactosamine (UDP-GalNAc) from UDP-N-acetylglucosamine (UDP-GlcNAc) in the presence of NAD+, an initial step in glycoprotein or glycolipid synthesis.[3]
Historical significance
[edit]Dr. Luis Leloir deduced the role of GALE in galactose metabolism during his tenure at the Instituto de Investigaciones Bioquímicas del Fundación Campomar, initially terming the enzyme waldenase.[4] Dr. Leloir was awarded the 1970 Nobel Prize in Chemistry for his discovery of sugar nucleotides and their role in the biosynthesis of carbohydrates.[5]
Structure
[edit]GALE belongs to the short-chain dehydrogenase/reductase (SDR) superfamily of proteins.[6] This family is characterized by a conserved Tyr-X-X-X-Lys motif necessary for enzymatic activity; one or more Rossmann fold scaffolds; and the ability to bind NAD+.[6]
Tertiary structure
[edit]GALE structure has been resolved for a number of species, including E. coli[7] and humans.[8] GALE exists as a homodimer in various species.[8]
While subunit size varies from 68 amino acids (Enterococcus faecalis) to 564 amino acids (Rhodococcus jostii), a majority of GALE subunits cluster near 330 amino acids in length.[6] Each subunit contains two distinct domains. An N-terminal domain contains a 7-stranded parallel β-pleated sheet flanked by α-helices.[1] Paired Rossmann folds within this domain allow GALE to tightly bind one NAD+ cofactor per subunit.[2] A 6-stranded β-sheet and 5 α-helices comprise GALE's C-terminal domain.[1] C-terminal residues bind UDP, such that the subunit is responsible for correctly positioning UDP-glucose or UDP-galactose for catalysis.[1]
Active site
[edit]The cleft between GALE's N- and C-terminal domains constitutes the enzyme's active site. A conserved Tyr-X-X-X Lys motif is necessary for GALE catalytic activity; in humans, this motif is represented by Tyr 157-Gly-Lys-Ser-Lys 161,[6] while E. coli GALE contains Tyr 149-Gly-Lys-Ser-Lys 153.[8] The size and shape of GALE's active site varies across species, allowing for variable GALE substrate specificity.[3] Additionally, the conformation of the active site within a species-specific GALE is malleable; for instance, a bulky UDP-GlcNAc 2' N-acetyl group is accommodated within the human GALE active site by the rotation of the Asn 207 carboxamide side chain.[3]
| Residue | Function |
|---|---|
| Ala 216, Phe 218 | Anchor uracil ring to enzyme. |
| Asp 295 | Interacts with ribose 2' hydroxyl group. |
| Asn 179, Arg 231, Arg 292 | Interact with UDP phosphate groups. |
| Tyr 299, Asn 179 | Interact with galactose 2' hydroxyl or glucose 6' hydroxyl group; properly position sugar within active site. |
| Tyr 177, Phe 178 | Interact with galactose 3' hydroxyl or glucose 6' hydroxyl group; properly position sugar within active site. |
| Lys 153 | Lowers pKa of Tyr 149, allows for abstraction or donation of a hydrogen atom to or from the sugar 4' hydroxyl group. |
| Tyr 149 | Abstracts or donates a hydrogen atom to or from the sugar 4' hydroxyl group, catalyzing formation of 4-ketopyranose intermediate. |
Mechanism
[edit]Conversion of UDP-galactose to UDP-glucose
[edit]GALE inverts the configuration of the 4' hydroxyl group of UDP-galactose through a series of 4 steps. Upon binding UDP-galactose, a conserved tyrosine residue in the active site abstracts a proton from the 4' hydroxyl group.[7][10]
Concomitantly, the 4' hydride is added to the si-face of NAD+, generating NADH and a 4-ketopyranose intermediate.[1] The 4-ketopyranose intermediate rotates 180° about the pyrophosphoryl linkage between the glycosyl oxygen and β-phosphorus atom, presenting the opposite face of the ketopyranose intermediate to NADH.[10] Hydride transfer from NADH to this opposite face inverts the stereochemistry of the 4' center. The conserved tyrosine residue then donates its proton, regenerating the 4' hydroxyl group.[1]
Conversion of UDP-GlcNAc to UDP-GalNAc
[edit]Human and some bacterial GALE isoforms reversibly catalyze the conversion of UDP-GlcNAc to UDP-GalNAc through an identical mechanism, inverting the stereochemical configuration at the sugar's 4' hydroxyl group.[3][11]
Biological function
[edit]
Galactose metabolism
[edit]No direct catabolic pathways exist for galactose metabolism. Galactose is therefore preferentially converted into glucose-1-phosphate, which may be shunted into glycolysis or the inositol synthesis pathway.[12]
GALE functions as one of four enzymes in the Leloir pathway of galactose conversion of glucose-1-phosphate. First, galactose mutarotase converts β-D-galactose to α-D-galactose.[1] Galactokinase then phosphorylates α-D-galactose at the 1' hydroxyl group, yielding galactose-1-phosphate.[1] In the third step, galactose-1-phosphate uridyltransferase catalyzes the reversible transfer of a UMP moiety from UDP-glucose to galactose-1-phosphate, generating UDP-galactose and glucose-1-phosphate.[1] In the final Leloir step, UDP-glucose is regenerated from UDP-galactose by GALE; UDP-glucose cycles back to the third step of the pathway.[1] As such, GALE regenerates a substrate necessary for continued Leloir pathway cycling.
The glucose-1-phosphate generated in step 3 of the Leloir pathway may be isomerized to glucose-6-phosphate by phosphoglucomutase. Glucose-6-phosphate readily enters glycolysis, leading to the production of ATP and pyruvate.[13] Furthermore, glucose-6-phosphate may be converted to inositol-1-phosphate by inositol-3-phosphate synthase, generating a precursor needed for inositol biosynthesis.[14]
UDP-GalNAc synthesis
[edit]Human and selected bacterial GALE isoforms bind UDP-GlcNAc, reversibly catalyzing its conversion to UDP-GalNAc. A family of glycosyltransferases known as UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosamine transferases (ppGaNTases) transfers GalNAc from UDP-GalNAc to glycoprotein serine and threonine residues.[15] ppGaNTase-mediated glycosylation regulates protein sorting,[16][17][18][19][20] ligand signaling,[21][22][23] resistance to proteolytic attack,[24][25] and represents the first committed step in mucin biosynthesis.[15]
Role in disease
[edit]Human GALE deficiency or dysfunction results in Type III galactosemia, which may exist in a mild (peripheral) or more severe (generalized) form.[12]
References
[edit]- ^ a b c d e f g h i j k Holden HM, Rayment I, Thoden JB (November 2003). "Structure and function of enzymes of the Leloir pathway for galactose metabolism". J. Biol. Chem. 278 (45): 43885–8. doi:10.1074/jbc.R300025200. PMID 12923184.
- ^ a b Liu Y, Vanhooke JL, Frey PA (June 1996). "UDP-galactose 4-epimerase: NAD+ content and a charge-transfer band associated with the substrate-induced conformational transition". Biochemistry. 35 (23): 7615–20. doi:10.1021/bi960102v. PMID 8652544.
- ^ a b c d Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM (May 2001). "Human UDP-galactose 4-epimerase. Accommodation of UDP-N-acetylglucosamine within the active site". J. Biol. Chem. 276 (18): 15131–6. doi:10.1074/jbc.M100220200. PMID 11279032.
- ^ LELOIR LF (September 1951). "The enzymatic transformation of uridine diphosphate glucose into a galactose derivative". Arch Biochem. 33 (2): 186–90. doi:10.1016/0003-9861(51)90096-3. hdl:11336/140700. PMID 14885999.
- ^ "The Nobel Prize in Chemistry 1970" (Press release). The Royal Swedish Academy of Science. 1970. Retrieved 2010-05-17.
- ^ a b c d Kavanagh KL, Jörnvall H, Persson B, Oppermann U (December 2008). "Medium- and short-chain dehydrogenase/reductase gene and protein families : the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes". Cell. Mol. Life Sci. 65 (24): 3895–906. doi:10.1007/s00018-008-8588-y. PMC 2792337. PMID 19011750.
- ^ a b PDB: 1EK5; Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM (May 2000). "Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase". Biochemistry. 39 (19): 5691–701. doi:10.1021/bi000215l. PMID 10801319.
- ^ a b c PDB: 1XEL; Thoden JB, Frey PA, Holden HM (April 1996). "Molecular structure of the NADH/UDP-glucose abortive complex of UDP-galactose 4-epimerase from Escherichia coli: implications for the catalytic mechanism". Biochemistry. 35 (16): 5137–44. doi:10.1021/bi9601114. PMID 8611497.
- ^ PDB: 1A9Z; Thoden JB, Holden HM (August 1998). "Dramatic differences in the binding of UDP-galactose and UDP-glucose to UDP-galactose 4-epimerase from Escherichia coli". Biochemistry. 37 (33): 11469–77. doi:10.1021/bi9808969. PMID 9708982.
- ^ a b Liu Y, Thoden JB, Kim J, Berger E, Gulick AM, Ruzicka FJ, Holden HM, Frey PA (September 1997). "Mechanistic roles of tyrosine 149 and serine 124 in UDP-galactose 4-epimerase from Escherichia coli". Biochemistry. 36 (35): 10675–84. doi:10.1021/bi970430a. PMID 9271498.
- ^ Kingsley DM, Kozarsky KF, Hobbie L, Krieger M (March 1986). "Reversible defects in O-linked glycosylation and LDL receptor expression in a UDP-Gal/UDP-GalNAc 4-epimerase deficient mutant". Cell. 44 (5): 749–59. doi:10.1016/0092-8674(86)90841-X. PMID 3948246. S2CID 28293937.
- ^ a b Lai K, Elsas LJ, Wierenga KJ (November 2009). "Galactose toxicity in animals". IUBMB Life. 61 (11): 1063–74. doi:10.1002/iub.262. PMC 2788023. PMID 19859980.
- ^ Stryer, Lubert; Berg, Jeremy Mark; Tymoczko, John L. (2008). Biochemistry (Looseleaf). San Francisco: W. H. Freeman. pp. 443–58. ISBN 9780716718437.
- ^ Michell RH (February 2008). "Inositol derivatives: evolution and functions". Nat. Rev. Mol. Cell Biol. 9 (2): 151–61. doi:10.1038/nrm2334. PMID 18216771. S2CID 3245927.
- ^ a b Ten Hagen KG, Fritz TA, Tabak LA (January 2003). "All in the family: the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases". Glycobiology. 13 (1): 1R – 16R. doi:10.1093/glycob/cwg007. PMID 12634319.
- ^ Alfalah M, Jacob R, Preuss U, Zimmer KP, Naim H, Naim HY (June 1999). "O-linked glycans mediate apical sorting of human intestinal sucrase-isomaltase through association with lipid rafts". Curr. Biol. 9 (11): 593–6. Bibcode:1999CBio....9..593A. doi:10.1016/S0960-9822(99)80263-2. PMID 10359703. S2CID 16866875.
- ^ Altschuler Y, Kinlough CL, Poland PA, Bruns JB, Apodaca G, Weisz OA, Hughey RP (March 2000). "Clathrin-mediated endocytosis of MUC1 is modulated by its glycosylation state". Mol. Biol. Cell. 11 (3): 819–31. doi:10.1091/mbc.11.3.819. PMC 14813. PMID 10712502.
- ^ Breuza L, Garcia M, Delgrossi MH, Le Bivic A (February 2002). "Role of the membrane-proximal O-glycosylation site in sorting of the human receptor for neurotrophins to the apical membrane of MDCK cells". Exp. Cell Res. 273 (2): 178–86. doi:10.1006/excr.2001.5442. PMID 11822873.
- ^ Naim HY, Joberty G, Alfalah M, Jacob R (June 1999). "Temporal association of the N- and O-linked glycosylation events and their implication in the polarized sorting of intestinal brush border sucrase-isomaltase, aminopeptidase N, and dipeptidyl peptidase IV". J. Biol. Chem. 274 (25): 17961–7. doi:10.1074/jbc.274.25.17961. PMID 10364244.
- ^ Zheng X, Sadler JE (March 2002). "Mucin-like domain of enteropeptidase directs apical targeting in Madin-Darby canine kidney cells". J. Biol. Chem. 277 (9): 6858–63. doi:10.1074/jbc.M109857200. PMID 11878264.
- ^ Hooper LV, Gordon JI (February 2001). "Glycans as legislators of host-microbial interactions: spanning the spectrum from symbiosis to pathogenicity". Glycobiology. 11 (2): 1R – 10R. doi:10.1093/glycob/11.2.1R. PMID 11287395.
- ^ Yeh JC, Hiraoka N, Petryniak B, Nakayama J, Ellies LG, Rabuka D, Hindsgaul O, Marth JD, Lowe JB, Fukuda M (June 2001). "Novel sulfated lymphocyte homing receptors and their control by a Core1 extension beta 1,3-N-acetylglucosaminyltransferase". Cell. 105 (7): 957–69. doi:10.1016/S0092-8674(01)00394-4. PMID 11439191. S2CID 18674112.
- ^ Somers WS, Tang J, Shaw GD, Camphausen RT (October 2000). "Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1". Cell. 103 (3): 467–79. doi:10.1016/S0092-8674(00)00138-0. PMID 11081633. S2CID 12719907.
- ^ Sauer J, Sigurskjold BW, Christensen U, Frandsen TP, Mirgorodskaya E, Harrison M, Roepstorff P, Svensson B (December 2000). "Glucoamylase: structure/function relationships, and protein engineering". Biochim. Biophys. Acta. 1543 (2): 275–293. doi:10.1016/s0167-4838(00)00232-6. PMID 11150611.
- ^ Garner B, Merry AH, Royle L, Harvey DJ, Rudd PM, Thillet J (June 2001). "Structural elucidation of the N- and O-glycans of human apolipoprotein(a): role of o-glycans in conferring protease resistance". J. Biol. Chem. 276 (25): 22200–8. doi:10.1074/jbc.M102150200. PMID 11294842.
Further reading
[edit]- Leloir LF (1953). "Enzymic Isomerization and Related Processes". Advances in Enzymology and Related Areas of Molecular Biology. Advances in Enzymology - and Related Areas of Molecular Biology. Vol. 14. pp. 193–218. doi:10.1002/9780470122594.ch6. ISBN 9780470122594. PMID 13057717.
{{cite book}}:|journal=ignored (help) - Maxwell ES, de Robichon-Szulmajster H (1960). "Purification of uridine diphosphate galactose-4-epimerase from yeast and the identification of protein-bound diphosphopyridine nucleotide". J. Biol. Chem. 235 (2): 308–312. doi:10.1016/S0021-9258(18)69520-1.
- Wilson DB, Hogness DS (August 1964). "The enzymes of the galactose operon in Escherichia coli. I Purification and characterization of uridine diphosphogalactose 4-epimerase". J. Biol. Chem. 239: 2469–81. doi:10.1016/S0021-9258(18)93876-7. PMID 14235524.
External links
[edit]- GeneReviews/NCBI/NIH/UW entry on Epimerase Deficiency Galactosemia
- OMIM entries on Epimerase Deficiency Galactosemia
- UDPgalactose+4-Epimerase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
