Murine respirovirus
| Murine respirovirus | |
|---|---|
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Negarnaviricota |
| Class: | Monjiviricetes |
| Order: | Mononegavirales |
| Family: | Paramyxoviridae |
| Genus: | Respirovirus |
| Species: | Murine respirovirus
|
| Synonyms | |
| |
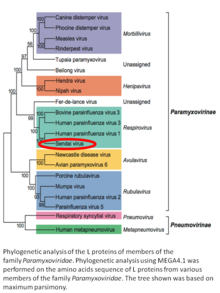
Murine respirovirus, formerly Sendai virus (SeV) and previously also known as murine parainfluenza virus type 1 or hemagglutinating virus of Japan (HVJ), is an enveloped, 150-200 nm–diameter, negative sense, single-stranded RNA virus of the family Paramyxoviridae.[2][3][4] It typically infects rodents and it is not pathogenic for humans or domestic animals.
Sendai virus (SeV) is a member of the genus Respirovirus.[5][6] The virus was isolated in the city of Sendai in Japan in the early 1950s. Since then, it has been actively used in research as a model pathogen. The virus is infectious for many cancer cell lines (see below), and has oncolytic properties demonstrated in animal models[7][8] and in naturally-occurring cancers in animals.[9] SeV's ability to fuse eukaryotic cells and to form syncytium was used to produce hybridoma cells capable of manufacturing monoclonal antibodies in large quantities.[10]
Recent applications of SeV-based vectors include the reprogramming of somatic cells into induced pluripotent stem cells[11][12] and vaccine creation. For vaccination purpose the Sendai virus-based constructs could be delivered in a form of nasal drops, which may be beneficial in inducing a mucosal immune response. SeV has several features that are important in a vector for a successful vaccine: the virus does not integrate into the host genome, it does not undergo genetic recombination, it replicates only in the cytoplasm without DNA intermediates or a nuclear phase and it does not cause any disease in humans or domestic animals. Sendai virus is used as a backbone for vaccine development against Mycobacterium tuberculosis that causes tuberculosis, against HIV-1 that causes AIDS and against other viruses, including those that cause severe respiratory infections in children.[13][14] The latter include Human Respiratory Syncytial Virus (HRSV), Human Metapneumovirus (HMPV) and Human Parainfluenza Viruses (HPIV).[14]
The vaccine studies against M. tuberculosis,[15] HMPV, HPIV1 and, HPIV2 are in the pre-clinical stage,[14] against HRSV a phase I clinical trail has been completed.[16] The phase I clinical studies of SeV-based vaccination were also completed for HPIV1.[14] They were done in adults and in 3- to 6-year-old children. As a result of vaccination against HPIV1 a significant boost in virus-specific neutralizing antibodies was observed.[14] A SeV-based vaccine development against HIV-1 has reached a phase II clinical trial.[17][18] In Japan intranasal Sendai virus-based SARS-CoV-2 vaccine was created and tested in a mouse model.[19]
As an infection agent
[edit]SeV replication occurs exclusively in the cytoplasm of the host cell. The virus is using its own RNA polymerase. One replication cycle takes approximately 12–15 hours with one cell yielding thousands of virions.[20]
Susceptible animals
[edit]The virus is responsible for a highly transmissible respiratory tract infection in mice, hamsters, guinea pigs, rats,[21] and occasionally marmosets,[22] with infection passing through both air and direct contact routes. Natural infection occurs by way of the respiratory tract. In animal facility airborne transmission can occur over a distance of 5–6 feet as well as through air handling systems. The virus can be detected in mouse colonies worldwide,[23] generally in suckling to young adult mice. A study in France reported antibodies to SeV in 17% of mouse colonies examined.[24] Epizootic infections of mice are usually associated with a high mortality rate, while enzootic disease patterns suggest that the virus is latent and can be cleared over the course of a year.[4] Sub-lethal exposure to SeV can promote long-lasting immunity to further lethal doses of SeV.[25] The virus is immunosuppressive and may predispose to secondary bacterial infections.[26] There are no scientific studies, which were performed using modern detection methods, which would identify SeV as an infectious and decease causative for humans or domestic animals.[16]


Variable susceptibility to infection in mouse and rat strains
[edit]Inbred and outbred mouse and rat strains have very different susceptibility to Sendai virus infection.[27] Visualization of SeV infection in live animals demonstrates this difference.[28] The 129/J mice tested were approximately 25,000-fold more sensitive than SJL/J mice.[29] C57BL/6 mice are highly resistant to the virus, while DBA/2J mice are sensitive.[30] C57BL/6 mice showed slight loss of body weight after SeV administration, which returned to normal later. Only 10% mortality rate was observed in C57BL/6 mice after the administration of very high virulent dose of 1*105 TCID50.[31] It was shown that resistance to the lethal effects of Sendai virus in mice is genetically controlled and expressed through control of viral replication within the first 72 hours of infection.[30] Treatment of both strains with exogenous IFN before and during viral infection led to an increase in survival time in C57BL/6 mice, but all animals of both strains ultimately succumb to SeV caused disease.[32] If a mouse survives a SeV infection, it develops a lifelong immunity to subsequent viral infections.[33]
There are SeV-resistant F344 rats and susceptible BN rats.[34]
Course of infection
[edit]In the host airways the virus titer reaches a peak after 5–6 days post infection initiation that decreases to undetectable levels by day 14.[35] The virus promotes a descending respiratory infection, which begins in the nasal passages, passes through the trachea into the lungs and causes necrosis of the respiratory epithelium. The necrosis is mild in the first few days of infection, but later became severe by peaking around day 5. By day 9, the cells of the surface of the airways regenerate. Focal interstitial pneumonia can developed accompanied by inflammation and lesions of various degrees on the lungs. Usually, the respiratory system shows signs of healing within 3 weeks of infection, however, residual lesions, inflammation, or permanent scarring can occur. 6–8 days after the infection initiation serum antibodies appear. They remain detectable for about 1 year.[35]
- Sneezing
- Hunched posture
- Respiratory distress
- Porphyrin discharge from eyes and/or nose
- Lethargy
- Failure to thrive in surviving babies and young rats
- Anorexia
Diagnosis and prophylaxis
[edit]SeV induces lesions within the respiratory tract, usually associated with bacterial inflammation of the trachea and lung (tracheitis and bronchopneumonia, respectively). However, the lesions are limited, and aren't indicative solely of SeV infection. Detection, therefore, makes use of SeV-specific antigens in several serological methods, including ELISA, immunofluorescence, and hemagglutination assays, with particular emphasis on use of the ELISA for its high sensitivity (unlike the hemagglutination assay) and its fairly early detection (unlike the immunofluorescence assay).[36]
In a natural setting, the respiratory infection of Sendai virus in mice is acute. From the extrapolation of the infection of laboratory mice, the presence of the virus may first be detected in the lungs 48 to 72 hours following exposure. As the virus replicates in the respiratory tract of an infected mouse, the concentration of the virus grows most quickly during the third day of infection. After that, the growth of the virus is slower but consistent. Typically, the peak concentration of the virus is on the sixth or seventh day, and rapid decline follows that by the ninth day. A fairly vigorous immune response mounted against the virus is the cause of this decline. The longest period of detected presence of the virus in a mouse lung is fourteen days after infection.[37]
Eaton et al. advises that, when controlling an outbreak of SeV, disinfecting the laboratory environment and vaccinating the breeders, as well as eliminating infected animals and screening incoming animals, should clear the problem very quickly. Imported animals should be vaccinated with SeV and placed in quarantine, while, in the laboratory environment, breeding programs should be discontinued, and the non-breeding adults isolated for two months.[38]
Virus induced immunosuppression
[edit]The virus is a powerful immunomodulator. SeV can act in both directions: it can activate or suppress the immune response depending on the type of cell, host and time period after infection initiation. The virus can suppress the IFN production and response pathways[39][40][41] as well as inflammation pathway.[42]
Apoptosis inhibition
[edit]Sendai virus P gene encodes a nested set of proteins (C', C, Y1 and Y2), which are named to collectively as the C proteins (see the section "genome structure" below). C proteins of SeV are able to suppress apoptosis.[43] The antiapoptotic activity of the C proteins supports SeV infection in the host cells.
Interferon production and signal transduction inhibition
[edit]The virus prevents the stimulation of type 1 IFN production and subsequent cell apoptosis in response to virus infection by inhibiting the activation of IRF-3.[39][40][41] Two virus proteins: C and V are mainly involved in this process. SeV can attenuate cell defense mechanisms and allow itself to escape from host innate immunity by inhibiting the interferon response pathway in addition to inhibiting the interferon production. The table below demonstrates the inhibition mechanism.
| protein | mapped to amino-acids | binding with | effect |
|---|---|---|---|
| C-protein | IKKα serine / threonine kinase | The binding prevents the phosphorylation of IRF7 and inhibits a pathway that includes a Toll-like receptor (TLR7) and TLR9-induction of IFN-alpha in plasmacytoid dendritic cells.[44] | |
| interferon-alpha/beta receptor subunit 2 (IFNAR2) | The binding inhibits IFN-α-stimulated tyrosine phosphorylation of the upstream receptor-associated kinases TYK2 and JAK1.[45] | ||
| 99-204[46] | N-terminal domain of STAT1 | The binding suppresses the signal transduction pathways of interferon alpha/beta (IFN-α/β) and IFN-γ[47][48] | |
| The C-protein inhibits the production of nitric oxide (NO) by murine macrophages[49][50] that has cytotoxic activity against viruses.[51] | |||
| The C protein decreases generation of dsRNA, therefore it is keeping PKR inactive to maintain proteins translation in a host cell.[52] | |||
| V-protein | MDA5 | The binding inhibits downstream MDA5 activation of the IFN promoter.[53][54] | |
| RIG-I | The binding prevents downstream RIG-I signaling to the mitochondrial antiviral signaling protein (MAVS) by disrupting TRIM25 -mediated ubiquitination of RIG-I.[55] The binding suppresses inducible NO synthase (iNOS) via the RIG-I/TRIM25 pathway and decreases the production of nitric oxide (NO) in infected macrophages.[56] | ||
| TRIM25 | |||
| The V-protein suppresses the production of interleukin-1β, by inhibiting the assembly of the inflammasome NLRP3.[57] | |||
| Y1-protein | 149-157[46] | Ran | The nuclear translocation of viral protein is mediated by Ran GTPase. The amino acids represent nuclear localization, which is believed to suppress IFN-induced gene expression.[46] |
Anti-IFN activity of C protein is shared across the family Paramyxoviridae, and therefore appears to play an important role in paramyxovirus immune evasion.[44] Human Parainfluenza Virus type 1 (HPIV1), which is a close relative of SeV and is (in contrast to SeV) a successful human pathogen, does not express V proteins, only C proteins. So, all needed functions provided by V in SeV can be provided by C in HPIV1. Thus, C and V have these "overlapping functions" because of the multi-faceted nature of host defense that can be countered at so many places, and exactly how well and where will in part explain host restriction.[58]
The C-protein also appears to be responsible for limiting the production of NO in infected macrophages, which in turn reduces inflammation. [59]
Host restriction and safety for domestic animals
[edit]Currently, there is no scientific data obtained using modern detection methods that would identify SeV as an infectious - disease causing agent for humans or domestic animals. Modern methods for the identification of pathogenic microorganisms have never detected SeV in pigs or other domestic animals, despite the isolation of other paramyxoviruses.[60][61][62][63] Consequently, it is recognized that Sendai virus disease causing infection is host restrictive for rodents and the virus does not cause disease in humans[64] or domestic animals, which are natural hosts for their own parainfluenza viruses. After experimental SeV infection the virus can replicate and shed from the upper and lower respiratory tract of African green monkeys and chimpanzees, but it is not causing any disease.[65] Sendai virus has been used and demonstrated high safety profile in clinical trials involving both adults[64] and children[66] to immunize against human parainfluenza virus type 1, since the two viruses share common antigenic determinants and trigger the generation of cross-reactive neutralizing antibodies. The study that was published in 2011 demonstrated that SeV neutralizing antibodies (which were formed due to human parainfluenza virus type 1 past infection) can be detected in 92.5% of human subjects worldwide with a median EC50 titer of 60.6 and values ranging from 5.9 to 11,324.[67] Low anti-SeV antibodies background does not block the ability of SeV-base vaccine to promote antigen-specific T cell immunity.[68]
Historic safety concerns
[edit]In 1952, Kuroya and his colleagues attempted to identify an infectious agent in human tissue samples at Tohoku University Hospital, Sendai, Japan. The samples were taken from the lung of a newborn child that was affected by fatal pneumonia. The primary isolate from the samples was passaged in mice and subsequently in embryonated eggs.[69][70] The isolated infectious agent was later called Sendai virus, which was used interchangeably with the name “Hemagglutinating Virus of Japan”. Kuroya and his colleagues were convinced that they isolated the virus, which is a new etiological agent for human respiratory infections. Later in 1954, Fukumi and his colleagues at the Japan National Institute of Health put forward an alternative explanation for the origin of the virus. It was suggested that the mice used to passage the virus were infected with the mouse virus. Thus, mouse virus was later transferred to embryonated eggs, isolated and finally named the Sendai virus.[71] This explanation of Fukumi, pointing to the mouse rather than the human origin of the virus, has been supported by numerous scientific data later. The historical aspects of the Sendai virus isolation and controversy behind it are well described in the review.[4] Thus, for some time, it was erroneously assumed that Sendai virus is human disease causing pathogen.[72] The incorrect assumption that the virus was isolated from human infectious material is still reported by Encyclopædia Britannica[73] and by ATCC in the description of the history of the viral isolate Sendai/52.[74] It was also believed that the virus could cause disease not only in humans but also in pigs, because antibodies to the virus were often found in their organisms during the swine epidemic in Japan in 1953–1956. High incidence of seropositivity to the virus was observed in pigs in 15 districts of Japan.[72] An explanation was later found for this widespread detection of antibodies (see the section below). Yet, despite overwhelming evidence that indicate that SeV is host restrictive rodent pathogen, in some veterinary manuals.[75] and safety leaflets,[76][77] SeV is still listed as a virus that can cause disease in pigs. Similar information is provided by Encyclopædia Britannica.[73] In reality, the multiple isolates of paramyxoviruses in pigs, using modern nucleic acid sequencing methods, have never been identified as SeV.[60][62][63]
Antigenic stability and cross-reactive antibodies
[edit]All viruses in the family Paramyxoviridae are antigenically stable; therefore the family representatives that are close relatives and belong to the same genus, most likely, share common antigenic determinants. Thus, porcine parainfluenza 1,[62][63] which has high sequence homology with SeV[62] and also belongs to the same genus Respirovirus as SeV, probably, has cross-reactive antibodies with SeV. Perhaps the porcine parainfluenza 1 was responsible for pigs disease in Japan in 1953–1956.[72] However, the antigenic cross-reactivity among these two representatives within the genus Respirovirus may explain why SeV antibodies were found in sick pigs, and why it was thought that SeV was the etiological causative agent of pigs disease. Human parainfluenza virus type 1, also shares common antigenic determinants with SeV and triggers the generation of cross-reactive neutralizing antibodies.[64] This fact can explain wide spread detection of SeV antibodies in humans in the 1950s-1960s.[72] Recently published study also showed this wide spread detection. The study that was published in 2011 demonstrated that SeV neutralizing antibodies (which were formed due to human parainfluenza virus type 1 past infection) can be detected in 92.5% of human subjects worldwide with a median EC50 titer of 60.6 and values ranging from 5.9 to 11,324.[67] Low anti-SeV antibodies background does not block the ability of SeV-base vaccine to promote antigen-specific T cell immunity.[68]
Virus shedding in airways of non-natural hosts
[edit]Sendai virus administration to non-natural hosts results in shedding virions in the airways. Thus, 10 hours later after intranasal SeV administration, infectious virions carrying foreign trans genes can be detected in sheep's’ lungs.[78] Moreover, SeV replicates to detectable levels in the upper and lower respiratory tract of African green monkeys and chimpanzees.[65]
Virus induced antiviral immunity
[edit]SeV can overcome antiviral mechanisms in some of its natural hosts (some rodents), but the virus is ineffective in overcoming these mechanisms in some other organisms that are virus resistant.[79] Both innate and adaptive immunity promote efficient recovery from SeV infection.[25]
SeV stimulates interferon production and transduction pathway
[edit]The main component of innate antiviral response is type I interferons (IFNs) production and most cells can produce type I IFNs, including IFN-α and -β.[80] The recognition by cellular molecules that are called pattern recognition receptors (PRR) of triggering viral elements, such as the virus genomic RNA, the replication intermediary double-stranded RNA, or the viral ribonucleoproteins, promotes IFN production and response pathways. Viral genomic and protein components can bind variable PRRs and stimulate a signaling pathway that results in the activation of the transcription factors, which relocate to nucleus and trigger type I IFNs transcription.

Interferon production
[edit]Because of powerful interferon stimulating properties, before recombinant interferon alpha became available for medical use, SeV was selected, among other viruses, for the industrial large-scale IFN production. A procedure involving inactivated SeV treatment of human peripheral blood leukocytes from donors’ blood was used for this production.[81]
Below is a table that listed known PRRs and interferon regulatory factors that are getting activated upon SeV infection.
| Molecules | Alias | Effect |
|---|---|---|
| Pattern recognition receptors (PRR) | ||
| Toll Like Receptors | TLR | SeV infection stimulates mRNA expression of TLR1, TLR2, TLR3, and TLR7 in macrophages. This effect is IFN-alpha/beta dependent because anti-IFN-alpha/beta neutralizing antibodies down regulate this mRNA transcription stimulation.[82] Human mast cell infection with SeV induces an antiviral response with activation of expression of type 1 IFN and TLR-3.[83] |
| NLRC5 | Using human embryonic kidney cells (HEK 293T) it has been shown that SeV can stimulate production of a pattern recognition receptor NLRC5, which is a cytosolic protein expressed mainly in hematopoietic cells.[84] | |
| Retinoic acid-inducible gene I | RIG-1 | RIG-1-IRF7-mediated induction of IFN-α by SeV requires both RIG-I and mitochondrial antiviral-signaling protein (MAVS) expression.[85] MAVS is also needed for SeV induction of IκB kinase (IKK), IRF3 and IFN-β in human cells.[85] Single-stranded Sendai virus genomic RNA bearing 5′-triphosphates activates the RIG-I mediated IFN-beta production.[86] SeV replication triggers activation of MAPK/ERK pathway (also known as the Ras-Raf-MEK-ERK pathway) in a RIG-I-dependent manner in dendritic cells (DC) and in fibroblasts. RIG-I-mediated activation of this pathway by SeV results in type I IFN production.[87] Human mast cell infection with SeV induces an antiviral response with activation of expression of type 1 IFN and RIG-1.[83] |
| Melanoma differentiation-associated antigen 5 | MDA5 | MDA5 has been shown to be an important participant in the antiviral SeV response and IFN type I production.[88] Human mast cell infection with SeV induces an antiviral response with activation of expression of type 1 IFN and MDA-5.[83] |
| Interferon regulatory factors | ||
| Interferon regulatory factor 3 | IRF-3 | SeV can activate the ubiquitously expressed IRF-3 by triggering its post-translational phosphorylation in human cells. IRF-3, is activated by phosphorylation on a specific serine residue, Ser396.[89] |
| Interferon regulatory factor 7 | IRF-7 | There is also some evidence that demonstrates that SeV activates IRF-7.[90] |
Many different cells can produce interferon in response to SeV
[edit]| Cell type | Effect |
|---|---|
| Human peripheral blood leukocytes | Sendai-virus-induced human peripheral blood leukocytes produce the interferon alpha (IFN-α)[91] and the interferon gamma (IFN-γ).[92][93] The SeV induced IFN-α consists from at least nine different sub types of IFN-α: 1a, 2b, 4b, 7a, 8b, 10a, 14c, 17b and 21b. Among these sub types IFN-α1 represents about 30% of total IFN-α.[94] It was shown that the HN of Paramyxoviruses is a potent inducer of type 1 IFN in human blood mononuclear cells.[95] |
| Lymphoid cells | SeV infection of Namalwa cells, which originated from human Burkitt lymphoma, transiently induces the transcriptional expression of multiple IFN-A genes.[96] It also has been shown that in these cells that SeV virus stimulates an expression of IFNα8, IFNα13, IFNβ and IFN type III (IFN-lambda, IFNλ): (L28α, IL28β, IL29).[97] |
| Monocytes and dendritic cells | Monocytes[98] and dendritic cells[99] produce IFN alpha/beta in response to SeV stimulation. However, plasmacytoid dendritic cells (pDC), despite inability to be infected by SeV,[100] produce higher level of IFN-1 compared to monocytes and monocyte-derived dendritic cells in response to SeV. This happens most likely due to the higher levels of constitutively expressed IRF-7 in pDC compared to monocytes and monocyte-derived dendritic cells.[101] The recognition of SeV by pDC happens through TLR7 activation and requires transport of cytosolic viral replication products into the lysosome by the process of autophagy. Moreover, for pDC, autophagy was found to be required for these cells production of IFN-α.[100]
Among conventional DCs,[102] only two subsets, namely CD4+ and CD8α− CD4− “double negative”[103] dendritic cells are capable of producing IFN-α and IFN-β in response to SeV infection. However, all conventional DC subsets, including CD8α+ can be infected with SeV.[104] SeV has the ability to replicate and to grow to high titers in human monocyte-derived DCs.[105] On the other hand, pDCs do not produce a significant number of SeV virions after infection.[100] When SeV is inactivated UV irradiation, it triggers lower levels of IFN-α production in pDCs compared to the levels induced by live virus.[100] Additionally, SeV has been shown to induce the production of IFN type III (IFN-lambda)[106] by human plasmacytoid dendritic cells.[107] In the case of mouse dendritic cells, UV-inactivated SeV can induce the production of type I IFN.[108] Similarly, some tumor cell lines also respond to UV-inactivated SeV by producing type I IFN.[109] However, similar to human pDCs, UV-inactivated SeV elicits lower levels of IFN-α production in mouse pDCs compared to the response triggered by live virus.[100] |
| Fibroblasts | Interferon-beta (IFN-β) production in human fibroblast cells also occurs in response to SeV treatment.[110] It has been shown that SeV infects human lung fibroblasts MRC-5 and induces the release of IFN-beta into the culture medium from these infected cells.[79] |
| Mast cells | Human mast cell infection SeV induces the expression of type 1 IFN.[83] |
| Astrocytes | SeV triggers high IFN-beta production in murine astrocytes.[111] This triggering is independent from TLR3 expression because it happens in TLR3 double negative mice.[111] |
| Spleen cells | HN of SeV can induce type 1 IFN production in mouse spleen.[112] |
Interferon response pathway protects some cells from SeV infection
[edit]SeV can stimulate and/or inhibit the IFN-beta response pathway depending on the type of cell and host. If SeV triggers IFN production, the produced IFN further protects cells from next rounds of SeV infection. Multiple examples of IFN-beta protecting cells from SeV are described. Pretreatment of human lung fibroblasts MRC-5 cells with IFN-beta inhibits the replication of SeV.[79]
A similar IFN-beta protection against the virus has been observed for some human malignant cells that maintain the IFN response pathway. HeLa cells can be infected with SeV; however, incubation of these cells with IFN-beta causes inhibition of SeV replication.[113] Multiple interferon stimulated genes (ISG) were identified as being required for this inhibition including IRF-9, TRIM69, NPIP, TDRD7, PNPT1 and so on.[113] One of this genes TDRD7 was investigated in more detail. The functional TDRD7 protein inhibits the replication of SeV and other paramyxoviruses, suppressing autophagy, which is necessary for productive infection with these viruses.[113]
SeV also triggers the expression of IFN induced Ifit2 protein that is involved in protecting mice from SeV through as yet unknown mechanism.[114] In addition, SeV triggers the expression of the chemokine interferon-γ inducible protein 10 kDa (CXCL10), which is involved in chemotaxis, induction of apoptosis, regulation of cell growth and mediation of angiostatic effects.[111] Human mast cell infection with SeV induces expression of interferon-stimulated genes MxA[115] and IFIT3[83] in addition to activation of expression of type 1 IFN, MDA-5, RIG-1 and TLR-3.
SeV stimulation of production of inflammatory cytokines, infammasomes and beta-defensins
[edit]Cytokines
[edit]Sendai virus can induce the production of many cytokines that enhance cellular immune responses. Some evidence that demonstrates that SeV activates the transcription factor NF-κB[116] and this activation helps in protection against SeV infection. SeV can stimulate the production of macrophage inflammatory protein-1α (MIB-1α) and –β (MIB-1β), RANTES (CCL5), tumor necrosis factor-alpha (TNF-alpha), tumor necrosis factor-beta (TNF-beta), interleukin-6 (IL-6 ), interleukin-8 (IL-8), interleukin-1 alpha (IL1A), interleukin-1 beta (IL1B), platelet-derived growth factor (PDGF-AB) and small concentrations of interleukin-2 (IL2) and GM-CSF.[93][92][91] Sendai virus can trigger production of interleukin 12 (IL12) and interleukin 23 (IL23) in human macrophages.[117] Even plasmids that deliver the F-coding gene of SeV to tumor cells in model animals trigger the production of RANTES (CCL5) in tumor-infiltrated T-lymphocytes.[108] SeV induces the production of B cell-activating factor by monocytes and by some other cells.[118] Heat-inactivated SeV virus induces the production of IL-10 and IL-6 cytokines by dendritic cells (DC).[119] Most likely, F protein is responsible for this induction because reconstituted liposomes containing F protein can stimulate IL-6 production by DC. The production of IL-6 in response to SeV infection is restricted to conventional dendritic cells (DCs]) subsets, such as CD4+ and double negative (dnDC).[104]
The UV-inactivated SeV (and likely the alive virus as well) can stimulate dendritic cells to secrete chemokines and cytokines such as interleukin-6, interferon-beta, chemokine (C-C motif) ligand 5, and chemokine (C-X-C motif) ligand 10. These molecules activate both CD8+ T cells as well as natural killer cells. UV-inactivated SeV triggers the production of an intercellular adhesion molecule -1 (ICAM-1, CD54), which is a glycoprotein that serves as a ligand for macrophage-1 antigen (Mac-1) and lymphocyte function-associated antigen 1 (LFA-1 (integrin)). This induced production happens through the activation of NF-κB downstream of the mitochondrial antiviral signaling pathway and the RIG-I. The increased concentration of ICAM-1 on the cells surface increases the vulnerability of these cells to natural killer cells.[120] It has been shown in the Namalwa cells that SeV virus stimulates an expression of many genes involved in immune defense pathways, such as type I and type II IFN signaling, as well as cytokine signaling. Among the ten most virus-induced mRNAs are IFNα8, IFNα13, IFNβ, IFNλ: (L28α, IL28β, IL29), OASL, CXCL10, CXCL11 and HERC5.[97]
Stimulation of inflammasome helps protect against SeV infection
[edit]Using human embryonic kidney cells (HEK 293T) it has been shown that SeV can stimulate production of a pattern recognition receptor NLRC5, which is a cytosolic protein expressed mainly in hematopoietic cells.[84] This production activates the cryopyrin (NALP3) inflammasome.[121] Using human monocytic cell line-1 (THP-1) it has been shown that SeV can activate signal transduction by mitochondrial antiviral-signaling protein signaling (MAVS), which is a mitochondria-associated adaptor molecule that is required for optimal NALP3-inflammasome activity. Through MAVS signaling SeV stimulates the oligomerization of NALP3 and triggers NALP3-dependent activation of caspase-1[122] that, in turn, stimulates caspase 1-dependent production of interleukine -1 beta (IL-1β).[123]
Stimulation of beta-defensin production
[edit]SeV is a very effective stimulant of expression of human beta-defensin-1 (hBD-1). This protein is a member of the beta-defensin family of proteins that bridges innate and adaptive immune responses to a pathogen infection.[124] In response to SeV infection, the production of hBD-1 mRNA and protein increases 2 hours after exposure to the virus in purified plasmacytoid dendritic cells or in PBMC.[125]
Long-term antiviral immunity
[edit]After viral infection in rodents, type I IFNs promote SeV clearance and speed up the migration and maturation of dendritic cells. However, soon after viral infection, animals efficiently generate cytotoxic T cells independently of type I IFN signaling and clear the virus from their lungs. Moreover, even the animals that are unresponsive to type I IFN develop long-term anti-SeV immunity in a form of memory response that includes generation of CD8+ T cells and neutralizing antibodies. This memory response can protect animals against further challenge with a lethal dose of virus.[25]
Phosphorylation
[edit]SeV infection causes changes in a host cell protein phosphorylation, triggering phosphorylation of at least of 1347 host proteins.[126]
As an oncolytic agent
[edit]Sendai virus-based anticancer therapy for model[7][8] and companion animals[9] has been reported in several scientific papers. The described studies demonstrate that Sendai virus has a potential of becoming a safe and effective therapeutic agent against a wide range of human cancers. High genomic stability of SeV is a very desirable trait for oncolytic viruses. SeV is not likely to evolve into a pathogenic strain or into a virus with decreased oncolytic potential. The cytoplasmic replication of the virus results in a lack of host genome integration and recombination, which makes SeV safer and more attractive candidate for broadly used therapeutic oncolysis compared to some DNA viruses or retroviruses.[127]
Safety for humans
[edit]One of the great advantages of the Sendai virus as a potential oncolytic agent is its safety. Even though the virus is widespread in rodent colonies[4] and has been used in laboratory research for decades,[128] it has never been observed that it can cause human disease. Moreover, Sendai virus has been used in clinical trials involving both adults[64] and children[66] to immunize against human parainfluenza virus type 1, since the two viruses share common antigenic determinants and trigger the generation of cross-reactive neutralizing antibodies.The Sendai virus administration in the form of nasal drops in doses ranging from 5 × 105 50% embryo infectious dose (EID50) to 5 × 107 EID50 induced the production of neutralizing antibodies to the human virus without any measurable side effects.The results of these trials represent additional evidence of Sendai virus safety for humans.The development of T cell-based AIDS vaccines using Sendai virus vectors reached phase II clinical trial. Evaluation of the safety and immunogenicity of an intranasally administered replication-competent Sendai Virus–vectored HIV Type 1 gag vaccine demonstrated: induction of potent T-Cell and antibody responses in prime-boost regimens.[18][17] Sendai virus also used as a backbone for vaccine against respiratory syncytial virus (RSV).[13][129]
Model cancers
[edit]For cancer studies, it is desirable that the oncolytic virus be non-pathogenic for experimental animals, but the Sendai virus can cause rodent disease, which is a problem for research strategies. Two approaches have been used to overcome this problem and make Sendai virus non-pathogenic for mice and rats. One of these approaches included the creation of a set of genetically modified attenuated viral strains. Representatives of this set were tested on model animals carrying a wide range of transplantable human tumors. It has been shown that they can cause suppression or even eradication of fibrosarcoma,[130][131] neuroblastoma,[132] hepatocellular carcinoma,[133] melanoma, squamous cell[134] and prostate carcinomas.[135] SeV construct suppresses micrometastasis of head and neck squamous cell carcinoma in an orthotopic nude mouse model.[136] Complete eradication of established gliosarcomas in immunocompetent rats has also been observed.[137] SeV constructs have also been created with a modified protease cleavage site in the F-protein. The modification allowed the recombinant virus to specifically infect cancer cells that expressed the corresponding proteases.[133][130]

Case 1. Male dog of 7 years old developed cutaneous, ulcerated, and poorly differentiated mastocytoma (35 mm diameter) located close to his anus. (1) Primary tumor; (2) 2 weeks after the first virus treatment; (3) 4 weeks after the first virus treatment.
Case 2. Male German shorthaired pointer of 9 years old developed subcutaneous, regional (stage 2) intermediately differentiated mastocytoma. The primary tumor was removed without clean margins. (1) secondary growth 1 week after the surgical procedure; (2) 2 weeks after the first virus treatment; (3) 5 weeks after the first virus treatment.
Another approach of making Sendai virus non-pathogenic included the short-term treatment of the virions with ultraviolet light. Such treatment causes a loss of the virus replication ability. However, even this replication-deficient virus can induce the cancer cells death and stimulate anti-tumor immunity. It can trigger extensive apoptosis of human glioblastoma cells in culture, and it can efficiently suppress the growth of these cells in model animals.[138] The ultraviolet light treated virus can also kill human prostate cancer cells in culture[139] by triggering their apoptosis and eradicate tumors that originated from these cells in immunodeficient model animals.[109] Moreover, it can stimulate immunomodulated tumor regression of colon[140] and kidney cancers[141][142] in immunocompetent mice. Similar regressions caused by the replication-deficient Sendai virus have been observed in animals with transplanted melanoma tumors.[143][144]
Natural cancers
[edit]Some cancer studies with non-rodent animals have been performed with the unmodified Sendai virus. Thus, after intratumoral injections of the virus, complete or partial remission of mast cell tumors (mastocytomas) was observed in dogs affected by this disease.[9] Short-term remission after an intravenous injection of SeV was described in a patient with acute leukemia treated in the Clinical Research Center of University Hospitals of Cleveland (USA) by multiple viruses in 1964.[145] It is also reported[8][146] that the Moscow strain of SeV[147] was tested by Dr. V. Senin[148] and his team as an anticancer agent in a few dozen patients affected by various malignancies with metastatic growth in Russia in the 1990s.[149] The virus was injected intradermally or intratumorally and it caused fever in less than half of the treated patients, which usually disappeared within 24 hours. Occasionally, the virus administration caused inflammation of the primary tumor and metastases. Clinical outcomes were variable. A small proportion of treated patients experienced pronounced long-term remission with the disappearance of primary tumors and metastases. Sometimes the remission lasted 5–10 years or more after virotherapy. Brief descriptions of the medical records of the patients that experiences long-term remission are presented in the patent.[149] Intratumoral injection of UV irradiated and inactivated SeV resulted in an antitumor effect in a few melanoma patients with stage IIIC or IV progressive disease with skin or lymph metastasis. Complete or partial responses were observed in approximately half of injected and noninjected target lesions.[150]
Anticancer mechanism
[edit]Direct cancer cells killing. Malignant cells are vulnerable to SeV infection.
[edit]Sendai virus can infect and kill variable cancer cells (see section Sensitive cell lines and virus strains). However, some malignant cells are resistant to SeV infection. There are multiple explanations for such resistance. Not all cancer cells have cell entry receptors for the virus and not all cancer cells express virus processing serine proteases. There are also other mechanisms that can make a cancer cell resistant to an oncolytic virus. For example, some cancer cells maintain interferon response system that completely or partially protects a host cells from a virus infection.[151] Therefore, biomarkers needed to be developed to identify tumors that might succumb to SeV mediated oncolysis.
Sendai virus cell entry receptors are often overexpressed in cancer cells.
[edit]SeV receptors are potential biomarkers for evaluation of the vulnerability of malignant cells to the virus. They represented by glycoproteins and glycolipids (see section "SeV cell entry receptors").The expression of some molecules that can facilitate SeV cell entry (see section “SeV cell entry receptors”), frequently, accelerates carcinogenesis and/or metastasis development. For example, the presence of Sialyl-Lewisx antigen (cluster of differentiation 15s (CD15s)), which is one of SeV cell entry receptors, on the outer cell membrane, correlates with invasion potential of malignant cells, tumor recurrence, and overall patient survival for an extremely wide range of cancers.[152][153] Therefore, SeV virus preferentially can enter such cells.
Metastatic cancer cells frequently express a high density of glycoproteins or glycolipids - molecules that are rich in sialic acid.[154] Expression of the Vim2 antigen, which is another SeV cell entry receptor, is very important for the extravascular infiltration process of acute myeloid leukemia cells.[155] GD1a,[156] ganglioside also serves as SeV receptor and is found in large quantities on the surfaces of breast cancer stem cells.[157] High cell surface expression of another SeV receptor - ganglioside sialosylparagloboside /SPG/ NeuAcα2-3PG.[158] characterizes lymphoid leukemia cells.[159][160] Among other receptors represented by gangliosides GT1b is highly expressed on the outer membranes of brain metastases cells that originate from an extremely broad range of cancer,[161] while GD1a,[156] GT1b[162] and GQ1b[163] can be detected in human gliosarcomas. However, their quantity is not exceeding the quantity in normal frontal cerebral cortex.[164] The asialoglycoprotein receptors that bind Sendai virus.[165][166] and serve as SeV cell entry receptors are highly expressed in liver cancers.[167][168]
| Receptors for SeV and their Expression in Malignancies | |||
|---|---|---|---|
| Receptor | Malignancy/effect of receptor expression | Reference | Monoclonal AB availability |
| Human asialoglyco-protein receptor 1 (ASFR1, ASGR1)[169] | High expression in liver cancer and occasionally moderate expression in gliomas, renal, pancreatic, colorectal, and ovarian cancers | [1] | Two variants [2] |
| Sialyl-Lewisx Antigen
(sLeX/CD15s) |
Non-small cell lung cancer/enhances post-operative recurrence | [170][171] | Many variants |
| Glioma cells, high grade glioma | [173][174] | ||
| Tumor-initiating cells in glioblastoma | [175] | ||
| Glioblastoma, astrocytoma, ependymoma | [176] | ||
| Malignant glio-neuronal tumors | [177] | ||
| Medulloblastomas | [178][179][180] | ||
| Lung cancer, distant metastases | [181] | ||
| Colorectal cancer/promotes liver metastases, decreases time of disease-free survival | [182][183][184] | ||
| Gastric cancers/decreases patient survival time | [185][186] | ||
| Breast cancer/decreases patient survival time | [187][188][189] | ||
| Prostate tumor/promotes bone metastases | [190][191][192] | ||
| Cell lines of variable origin/high expression enhances adhesion of malignant cells to vascular endothelium | [193] | ||
| Variable cancers/high expression related to lymphatic invasion, venous invasion, T stage, N stage, M stage, tumor stage, recurrence, and overall patient survival | Review[194] | ||
| VIM-2 antigen
(CD65s) |
Acute myeloblastic leukemias | [195][196][197] | |
| GD1a | Breast cancer stem cells | [198] | |
| Glioma, glioblastoma | [173][199] | ||
| Castration resistant prostate cancer cells | [200] | ||
| GT1b | Brain metastases from colon, renal, lung, esophagus, pancreas, and mammary carcinomas | [201] | |
| SPG | Castration resistant prostate cancer cells | [200] | One variant |
| Lymphoid leukemia cells | [203][202] | ||
Cellular expression of glycoproteins can be evaluated by various molecular biology methods, which include RNA and protein measurements. However, cellular expression of gangliosides, which are sialic acid-containing glycosphingolipids, cannot be evaluated by these methods. Instead, it can be measured using anti-glycan antibodies, and despite the large collection of such antibodies in a community resource database, they are not always available for each ganglioside.[204] Therefore, indirect measurement of ganglioside expression by quantifying the levels of fucosyltransferases and glycosyltransferases that complete glycan synthesis is an alternative. There is evidence that expression of these enzymes and the production of gangliosides strongly correlate.[160] At least four representatives of fucosyltransferases and several glycosyltransferases including sialyltransferases are responsible for the synthesis of gangliosides that can serve as SeV receptors. All these proteins are often overexpressed in various tumors, and their expression levels correlate with the metastatic status of the tumor and the shorter life span of the patients. Thus, these enzymes are also potential biomarkers of SeV-oncolytic infectivity
| Synthesizing enzymes for SeV cell entry receptors | ||
| SeV receptor | Type of enzyme | Enzyme |
| Sialyl-Lewisx antigen/(sLeX/CD11s)[205][206][207][208] | Fucosyltransferase | FUT3, FUT5, FUT6, FUT7 |
| Glycosyltransferase | ST3GAL3, [3] [4] ST3GAL4,[5] [6] ST3GAL6 [7] [8] [9] | |
| Vim2 antigen /(CD65)[207] | Fucosyltransferase | FUT5 |
| GD1a[209][210][160][211] | Glycosyltransferases | ST3GAL1,[10] [11] ST3GAL2,[12] [13] ST6GALNAC5 [14] [15] ST6GALNAC6 [16] [17] |
| GD1b,[210] (GT1a, GQ1b and GP1c)[209] | ST6GALNAC6 [18] [19] | |
| GT1b[212] | ST3GAL2,[20] [21] | |
| Sialosylparagloboside (SPG).[160] | ST3GAL6, [22] [23] | |
Sendai virus proteolytic processing enzymes are often overexpressed in cancer cells.
[edit]The fusion protein (F) of SeV is synthesized as an inactive precursor and is activated by proteolytic cleavage of the host cell serine proteases (see the section “Proteolytic cleavage by cellular proteases” below). Some of these proteases are overexpressed in malignant neoplasms. For example, transmembrane serine protease 2 (TMPRSS2), which is an F-protein-processing enzyme, is often overexpressed in prostate cancer cells.[213] It is also overexpressed in some cell lines originating from various malignant neoplasms. Thus, it is highly expressed in bladder carcinoma,[214] human colon carcinoma CaCo2[215] and breast carcinomas SK-BR-3, MCF7 and T-47d.[216] TMPRSS2 is overexpressed in cervical and endocervical squamous cell carcinomas, along with colon, prostate, and rectum adenocarcinomas.[217] It is also overexpressed in uterine corpus endometrial and uterine carcinosarcomas.[217] Another F-protein-protease is tryptase beta 2 (TPSB2). This protease (with alias such as tryptase-Clara and mast cell tryptase) is expressed in normal club cells and mast cells, and in some cancers.[218] It's especially high expression is observed in the human mast cell line HMC-1,[219][220] and in the human erythroleukemia cell line HEL.[221][219] The release of this tryptase from mast cells enhances tumor cell metastasis.[222] Plasminogen (PLG), from which originates the mini-plasmin that can cleave the F-protein, is highly expressed in liver cancers.[223] Its expression is also increased in a wide range of other malignant neoplasms.[223] Factor X (F10) is frequently expressed in normal liver and in liver cancers.[224] SeV constructs were created with a modified protease cleavage site. The modification allowed the recombinant virus to specifically infect cancer cells that expressed the corresponding proteases, which can cleave a modified protease cleavage site.[130][133]
Defects in the interferon system
[edit]The interferon production and / or response system often malfunctions in malignant cells; therefore, they are much more vulnerable to infection with oncolytic viruses compared to normal cells[151] Thus, cells belonging to three human cell lines, originated from variable malignancies, such as U937, Namalwa, and A549, retain their ability to become infected with SeV even after treatment with type 1 IFN. Interferon response system is broken in these cells and it cannot protect them from SeV infection.[225]
In Namalwa cells SeV virus stimulates an expression of many genes involved in immune defense pathways, such as type I and type II IFN signaling, as well as cytokine signaling. Among the ten most virus-induced mRNAs are IFNα8, IFNα13, IFNβ, IFNλ: (L28α, IL28β, IL29), OASL, CXCL10, CXCL11 and HERC5.[97] However, despite stimulation of these genes expression by SeV, Namalwa cells can't protect themselves from the virus infection.
Ability of Sendai virus to inhibit interferon response in some cancer cells
[edit]In HeLa cells SeV (in contrast to Vesicular Stomatitis Virus) can counteract IFN-α pretreatment and keep a viral protein translation level similar to that in IFN-untreated cells.[52]
Activation of a necroptotic pathway in malignant cells
[edit]It has been shown, using fibrosarcoma cell line L929, that SeV is able to induce malignant cell death through necroptosis.[226] This type of cell death is highly immunogenic because dying necroptotic cells release damage-associated molecular pattern (DAMPs) molecules, which initiate adaptive immunity. The necroptotic pathway, triggered by SeV, requires RIG-I activation and the presence of SeV encoded proteins Y1 and/or Y2.[226]
Removing sialic acid residues from T-regulatory cell surfaces
[edit]Viral neuraminidase has the ability to remove sialic acid residues from cell surfaces,[227] including those on T-regulatory (Treg) cells. Research indicates that the Sialyl-Lewis x antigen is specifically found in activated, terminally differentiated, and highly suppressive CD4+ regulatory T (Treg) cells, which can be distinguished from nonsuppressive T cell.[228][229] Removing the suppressive Treg cells from human blood has been shown to enhance immune responses against tumor and viral antigens in vitro.[228] Removing the Sialyl-Lewis x antigen from Treg cells can inactivate their suppressive function.[229]
Virus, mediated fusion of cancer cells, kills them faster
[edit]The host organism fights viral infection using various strategies. One such strategy is the production of neutralizing antibodies. In response to this production, viruses have developed their own strategies for spreading the infection and avoiding the inactivation by the host produced neutralizing antibodies. Some viruses, and in particular paramyxoviruses, can produce new virus particles by fusing infected and healthy host cells. This fusion leads to the formation of a large multi-nuclear structure (syncytium). Sendai virus, as a representative of Paramyxoviridae, uses this strategy to spread its infection (see the section “Directed cell fusion” below). The virus can fuse up to 50-100 cells adjacent to one primary infected cell. This multi-nuclear formation, derived from several dozens of cells, survives for several days and subsequently releases functional viral particles.[8]
It has been demonstrated that the ability of a virus to destroy tumor cells increases along with an increase in the ability of the virus to form large multi-nuclear structures. The transfer of genes that are responsible for the formation of syncytium from the representative of Paramyxoviridae to the representatives of Rhabdoviridae or Herpesviridae makes the recipient viruses more oncolytic.[230][231] Moreover, the oncolytic potential of paramyxovirus can be enhanced by mutations in the fusion (F) gene protease-cleavage site, which allows the F-protein to be more efficiently processed by cellular proteases.[232] The introduction of the F gene of SeV in the form of a plasmid into the tumor tissue in mice by electroporation showed that the expression of the F gene increases the T cell infiltration of the tumor with CD4 + and CD8 + cells and inhibits tumor growth.[233] It was also shown in other similar experiments that cancer cells themselves, transfected with plasmids that encode viral membrane glycoproteins with fusion function, cause the collective death of neighboring cells forming syncytium with them. Recruitment of bystander cells into the syncytium leads to significant regression of the tumor.[234][235][236]
Killing of malignant cells by virus triggered anti-tumor immunity
[edit]The virus triggers indirect immunomodulated death of malignant cells using a number of mechanisms, which are described in a published review.[8] The viral enzyme neuraminidase (NA), which has sialidase activity, can make cancer cells more visible to the immune system by removing sialic acid residues from the surface of malignant cells.[8] SeV activates natural killer cells (NK), cytotoxic T lymphocytes (CTL) and dendritic cells (DC). The secretion of interleukin-6, that is triggered by the virus, also inhibits regulatory T cells.[140][108]
Stimulation of the secretion of cytokines
[edit]
Interferons
[edit]Type I and type II interferons have anticancer activity (see the "Function" section in the "Interferon" article). Interferons can promote expression of major histocompatibility complex molecules, MHC I and MHC II, and stimulate immunoproteasome activity. All interferons drastically increase the presentation of MHC I dependent antigens. Interferon gamma (IFN-gamma) also strongly promotes the MHC II-dependent presentation of antigens.[237] Higher MHC I expression leads to higher presentation of viral and abnormal peptides from cancer cells to cytotoxic T cells, while the immunoproteasome more efficiently processes these peptides for loading onto the MHC I molecule. Therefore, the recognition and killing of infected or malignant cells increases. Higher MHC II expression enhances presentation of viral and cancer peptides to helper T cells; which are releasing cytokines (such as more interferons, interleukins and other cytokines) that stimulate and co-ordinate the activity of other immune cells.[238][239][240]
By down regulation of angiogenic stimuli produced by tumor cells interferon can also suppress angiogenesis[241] In addition, they suppress the proliferation of endothelial cells. Such suppression causes a decrease in tumor vascularization and subsequent growth inhibition. Interferons can directly activate immune cells including macrophages and natural killer cells.[238] INF-1 and interferon gamma (IFN-γ) production are triggered by SeV molecular components in many cells (See "Virus induced antiviral immunity" section above).[91][92][93][110] It has been demonstrated that SeV can also induce the production of IFN type III (IFN-lambda)[106] by human plasmacytoid dendritic cells.[107]
Non interferons
[edit]Sendai virus can induce the production of many cytokines that enhance cellular immune responses against cancer cells. SeV stimulates the production of macrophage inflammatory protein-1α (MIB-1α) and –β (MIB-1β), RANTES (CCL5), tumor necrosis factor-alpha (TNF-alpha), tumor necrosis factor-beta (TNF-beta), interleukin-6 (IL-6 ), interleukin-8 (IL-8), interleukin-1 alpha (IL1A), interleukin-1 beta (IL1B), platelet-derived growth factor (PDGF-AB) and small concentrations of interleukin-2 (IL2) and GM-CSF.[93][92][91] The virus can trigger production of interleukin 12 (IL12) and interleukin 23 (IL23) in human macrophages.[117] Even plasmids that deliver the F-coding gene of SeV to tumor cells in model animals trigger the production of RANTES (CCL5) in tumor-infiltrated T-lymphocytes.[108]
SeV induces the production of B cell-activating factor by monocytes and by some other cells.[118]
Heat-inactivated SeV virus induces the production of IL-10 and IL-6 cytokines by dendritic cells (DC).[119] Most likely, F protein is responsible for this induction because reconstituted liposomes containing F protein can stimulate IL-6 production by DC. The production of IL-6 in response to SeV infection is restricted to conventional dendritic cells (DCs) subsets, such as CD4+ and double negative (dnDC).[104]
The UV-inactivated SeV (and likely the alive virus as well) can stimulate dendritic cells to secrete chemokines and cytokines such as interleukin-6, interferon-beta, chemokine (C-C motif) ligand 5, and chemokine (C-X-C motif) ligand 10. These molecules activate both CD8+ T cells as well as natural killer cells and attract them to the tumor. It has been shown that in cancer cell lines, UV-inactivated SeV triggers the production of an intercellular adhesion molecule -1 (ICAM-1, CD54), which is a glycoprotein that serves as a ligand for macrophage-1 antigen (Mac-1) and lymphocyte function-associated antigen 1 (LFA-1 (integrin)). Mac-1 and LFA-1 are receptors found on leukocytes. This induced production happens through the activation of nuclear factor-κB downstream of the mitochondrial antiviral signaling pathway and the retinoic acid-inducible gene I. The increased concentration of ICAM-1 on the surface of cancer cells, which is triggered by SeV, increases the vulnerability of these cells to natural killer cells.[120]
Neuraminidase (NA) removal of sialic acid from the surface of malignant cells stimulates natural killers cells and cytotoxic T lymphocytes
[edit]Increased sialylation levels on the cell membrane have been linked to a heightened potential for invasion and metastasis in cancer cells. This correlation has been observed across various models, including murine,[242][243][244] rat,[245] and human,[246] and is associated with the progression of malignancy. Some sialylation inhibitors can make cancer cells less malignant.[247][248][249]
One possible explanation for the relationship between increased sialylation and a malignant phenotype is that sialylation results in a thick layer of coating on the cell membrane that masks cancer antigens and protects malignant cells from immune surveillance. The activity and cytotoxicity of NK cells is inhibited by the expression of sialic acids on the tumor cell surface.[250] Removal of sialic acid residues from the surface of tumor cells makes them available to NK cells and cytotoxic T lymphocytes and, therefore, reduces their growth potential. Moreover, treating tumor cells with sialidase improves activation of NK cell secretion of IFN-γ.[250]
Some paramyxoviruses, including SeV encode and synthesize neuraminidase (sialidase), which can remove sialic acid residues from the surface of malignant cells. Hemagglutinin-neuraminidase (HN) is a single protein that induces hemagglutination and possesses neuraminidase (sialidase) activity. Neuraminidase (NA), a subunit of the HN protein, binds to and cleaves sialic acid from the cell surface.[251] NA also promotes cell fusion, which helps the nascent virions to avoid contact with host antibodies and thus enables the virus to spread within tissues.
Sialidase treatment of cells causes loss of sialic acid residues. This loss significantly increases the ability of malignant cells to activate cytotoxic T lymphocytes.[252] Variable sialidases can cause this effect,[252] including NA from Newcastle disease virus that have been shown to cleave 2,3-, 2,6-,[253] and 2,8-linkages between sialic acid residues.[254] In vitro, there was no significant difference between NAs from Newcastle disease virus, SeV and mumps virus[255] with respect to substrate specificity. These results suggest that treating a tumor with the virus results in desialylation of malignant cells, which contributes to increased anti-tumor immune surveillance. Therefore, the ability of SeV sialidase (NA) to remove sialic acid from the surface of malignant cells most likely helps to ensure the availability of tumor antigens for recognition by cytotoxic T lymphocytes.[252]
Stimulation of natural killer (NK) cells
[edit]Experiments with UV-inactivated SeV showed that NK cells are important in virus-mediated inhibition of tumor growth. This was shown in a mouse model of renal cancer, in which the anti-tumor effect of SeV was suppressed by reducing the number of NK cells by co-injection of specific antibodies.[141]
The activation of NK requires several receptors, among which are natural killer proteins 46 (NKp46) and 44 (NKp44). Studies have shown that the only paramyxovirus protein that activates NK is HN.[256] HN protein binding to NKp46 and/or NKp44 results in the lysis of cells whose surfaces display the HN protein or its fragments.[257][258] It can be assumed that NK activation and tumor suppression by UV-treated SeV[141] are caused by interaction between HN belonging to SeV, and NKp46 and/or NKp44 receptors belonging to NK cells.
Induction of anti-tumor cytotoxicity of cytotoxic T cells
[edit]SeV even after UV inactivation, being injected intratumorally, can cause tumor infiltration by dendritic cells (DCs) and CD4+ and CD8+ T, and it also can cause enhancing of anti-tumor activity of these cells.[140] Most likely, viral hemagglutinin-neuraminidase protein, highly contributes to the effect (see "Neuraminidase (NA) removal of sialic acid from the surface of malignant cells stimulates natural killers cells and cytotoxic T lymphocytes" section above).This hypothesis is based on two observations. First, the functional hemagglutinin-neuraminidase protein of the oncolytic Newcastle disease virus (NDV), which is a relative of SeV, has been shown to enhance the tumor-specific cytotoxic response of CD8+ T-cells and to increase the activity of CD4+ T-helper cells.[258] Second, UV-inactivated NDV, which is can not replicate, promotes anti-tumor CTL response as well as does intact NDV, which can replicate.[258] Since the hemagglutinin-neuraminidase proteins of the SeV and NDV viruses are highly homologous and function similarly,[255] it is likely that the HN protein of the SeV virus can activate both CTL and natural killers cell responses. Most likely neuraminidase removal of sialic acid from the surface of malignant cells contributes to this effects.[252]
SeV stimulation of dendritic cells
[edit]UV-inactivated SeV can cause dendritic cells (DCs) to maturate and to infiltrate a tumor.[140] Ex vivo infection of DCs with recombinant non-transmissible SeV induces maturation and activation of DCs[259] within 60 minutes.[260] When activated DCs that carry non-transmissible variants of SeV are administered, survival of animals injected with malignant melanoma,[261][262] colorectal cancer,[263] squamous cell carcinoma,[264] hepatic cancer, neuroblastoma, and prostate cancer[135] is significantly improved. It has been shown that the administration of such DCs prior to tumor cell injection prevents metastasis of neuroblastoma and prostate adenocarcinoma to the lungs.[265][266]
The enzymatic removal of sialic acids from the surface of dendritic cells by sialidase significantly promote the antigen-induced activation of naive T cells, while concurrently enhancing the resurgence of effector T cells. It is plausible that sialidase from Sendai virus (SeV) could execute this function.[267] The removal not only improves antigen cross-presentation but boosts anti-tumor immune responses as well.[268] Dendritic cells with reduced sialylation form higher avidity interactions with CD8+ T cells.[269]
SeV can replicate to high titers in human monocyte-derived DCs.[105][270] With the multiplicity of infection of 2, approximately 1/3 of the DCs begin to express encoded SeV proteins 8 hours after infection. This proportion increases to 2/3, 24 hours and decreases to 1/3, 48 hours after infection. SeV demonstrates high cytopathic effect on DCs; the virus can kill a third of DC even with a very low multiplicity of infection such as 0.5. Most important observation is that SeV infection triggers DC maturation, which is manifested in DC cell surface markers composition. The virus increases the expression of class I and class II molecules of the major histocompatibility complex (MHC) (HLA-A, HLA-B, HLA-C and HLADR), CD83, as well as costimulatory molecules CD40 and CD86.[270]

SeV suppression of regulatory T cells
[edit]Experiments with animal models have shown that, even after UV inactivation, SeV can block T-cell-mediated regulatory immunosuppression in tumors. The blocking mechanism is associated with the stimulation of SeV inactivated virions of interleukin 6 (IL-6) secretion by mature DCs. These effects lead to the eradication of most model tumors and inhibit the growth of the rest.[140] It has been shown that F protein alone can trigger IL-6 production in DC in a fusion-independent manner.[108]
As a vector
[edit]


SeV has been known to the research community since the late 1950s and has been widely used to create numerous variants of genetically engineered constructs, including vectors for transgene delivery.[271][128][272] Creation of SeV genetic constructs is easier compared to other viruses, many SeV genes have a transcriptional initiation and termination signals. Therefore, constructing a recombinant virus is straightforward; the foreign gene can be introduced into the viral genome by replacing or adding viral protein expressing gene(s). SeV can include a foreign gene or even multiple genes of large size. It has been demonstrated that a gene of more than 3 kb can be inserted and expressed in SeV.[273] Due to exclusively cytoplasmic replication, the virus does not carry the risk of genetic integration into the host genomes, which is a problem for many other viral vectors. The genome of SeV as genomes of other non segmented negative-stranded RNA viruses[274][275] has a low rate of homologous recombination and evolves comparatively slowly. Multiple reasons for this genomic stability exist: (1) the genome is nonsegmented, therefore cannot undergo genetic reassortment, (2) each protein and each amino acid has an important function. Therefore, any new genetic insertion, substitution or deletion would lead to a decrease or total loss of function that would in turn cause the new virus variant to be less viable. (3) Sendai virus belongs to a category of viruses that are governed by the “rule of six”.[276] SeV genome as genomes of other paramyxoviruses mainly include six genes, which encode for six major proteins. Low rate of homologous RNA recombination in paramyxoviruses probably results from this unusual genomic requirement for polyhexameric length (6n+0). Natural high genomic stability of SeV is a positive feature for it potential use as a vaccine vector or as an oncolytic agent. For any clinical or industrial applications, it is important that SeV genomic and inserted foreign genes would be expressed in a stable way. Due to SeV genetic stability, multiple serial passages of the virus construct in cell cultures or embryonated chicken eggs without drastic genomic changes are possible.[127] SeV constructs are known to stably express a wide variety of heterologous antigens.[277][127][278]
Reverse genetic system
[edit]The reverse genetics system to rescue Sendai virus was created and published in 1995.[279] Since then a number of modifications and improvements were described for representatives of Mononegavirales,[280] Paramyxoviridae in general,[281][282][283] and for Sendai virus in particular.[284] The entire length of the vector SeV genome, including transgenes, has to be arranged in multiples of six nucleotides (the so-called "rule of six").[276]
Genes addition, deletion and modification
[edit]Recombinant SeV variants has been constructed by introducing new genes and/or by deleting some viral genes such as F, M, and HN from the SeV genome.[263][285][286] Reporter genes, such as those that are coding luciferase,[28][287][288] green[289][290][291] or red[292] fluorescent proteins can be inserted in different locations in the viral genome. These locations include positions upstream of the N gene,[290][291][288] between the N and P genes,[293][292] between P and M,[28][287] M and F,[28][287][294][295] F and HN,[28][287] HN and L,[295] and after the L gene.[289]
SeV constructs have also been created with a modified protease cleavage site in fusion protein (F).[130][133][296][297] The SeV F protein is a type I membrane glycoprotein that is synthesized as an inactive precursor (F0) that must be activated by proteolytic cleavage at residue arginine-116.[4] After the cleavage F0 precursor yields two disulfide-linked subunits F1 and F2.[298] The proteolytic cleavage site can be changed, so other host proteases would be capable to process F0.[130][133][296][297]
Sendai virus based vector system that can deliver CRISPR/Cas9 for efficient gene editing was created.[299]
Non-invasive imaging
[edit]A variety of Sendai virus constructs carrying reporter genes were developed for non-invasive imaging of the virus infection in animals. They allow to study dynamics of SeV spread and clearance.[28][287] Some of these constructs were designed to deliver luciferase genes,[28][287][288] some to deliver green fluorescent protein (GFP),[290][291][294] others to deliver red fluorescent protein (RFP).[292]
Sendai virus minigenome
[edit]
The Sendai virus minigenome is a shortened version of its viral genome, in which some portions of the coding sequences of the virus have been removed. The removed genes can be replaced with a foreign trans-gene of interest.[300] The minigenome can be multiplied in cells expressing a minimal set of complementary viral proteins or infected with a homologous wild-type helper virus. Sendai virus minigenomes are used to produce recombinant proteins of interest,[300] and in a vector system to reprogram cells into pluripotent stem cells (iPSCs).[11][12] A Sendai virus minigenome lacking Fusion protein, but expressing Emerald Green Fluorescent Protein (EmGFP) as a reporter was found to be an efficient gene delivery vector in several human pancreatic cancer cells.[301]
Insertion of trans-genes into Sendai virus minigenome
[edit]To integrate a gene fragment of interest into the Sendai Virus genome, the following protocol[11] might be used.
The amplified gene fragment is inserted into a Sendai Virus vector lacking the F protein (SeV/ΔF). The recovery and amplification of SeV/ΔF vectors proceed as follows:
- Transfection: 293T cells are transfected with the pSeV/ΔF template containing the transgene of interest, along with plasmids that encode the T7 RNA polymerase and the viral genes NP, P, F5R (a modified F protein), and L.
- Cultivation: Post-transfection, the cells are incubated and cultured for 1 to 3 days to produce the initial SeV/ΔF vector.
- Propagation: The vector is then propagated in LLC-MK2/F7/A cells, a specialized cell line of LLC-MK2 that expresses the Sendai virus F protein, in a medium that includes trypsin.
- Titer Quantification: The titers of the harvested SeV vector are determined by measuring the cell infectious units (CIU) per milliliter through immunostaining with anti-SeV rabbit polyclonal serum.
Production of foreign soluble glycoproteins using Sendai Virus minigenome and allantoic fluid eхpression system
[edit]
Researchers have developed an innovative method to efficiently produce substantial quantities of heterologous viral glycoproteins within the allantoic cavity of embryonated chicken eggs.[300] This technique utilizes a Sendai virus minigenome as a vector to express soluble variants of specific proteins, notably the human respiratory syncytial virus (HRSV) and human metapneumovirus (HMPV) fusion (F) proteins. These proteins are engineered without their transmembrane and cytoplasmic domains, facilitating their solubility.
Overview of the Methodology:
- Rescue of Sendai Virus Minigenomes: The process begins with the rescue of Sendai virus minigenomes that encode the target proteins. This step is conducted in cell cultures, assisted by the wild-type Sendai virus, which acts as a helper virus.
- Propagation in Embryonated Eggs: Subsequently, the engineered viruses are propagated within the allantoic cavity of chicken embryonated eggs. This environment is conducive to the viruses’ replication and protein production. To enhance yields, the process may undergo several iterations across different generations of eggs.
- Yield Enhancement: Comparative studies have demonstrated that this method yields protein quantities that are 5 to 10 times greater than those produced in cell culture supernatants infected with vaccinia virus recombinants.

One of the latest applications of SeV-based vectors is the reprogramming of somatic cells into induced pluripotent stem cells.[11][12] The SeV vector with a mutation that is responsible for temperature-sensitive phenotype was created to facilitate the erasure of the vector genome in a cell line.[12] Temperature sensitive mutants of SeV encoding human OCT3/4, SOX2, KLF4 and c-MYC genes are used to infect human donor cells, but the resulting iPSCs became trans-gene free.[302] One possible source of donor cells are human cord blood-derived hematopoietic stem cells stimulated with cytokines. Among these cells SeV achieves high transgene expression in CD34+ cells subset.[303] Another source—human primary PBMC, according to a technical note of TaKaRa human primary PBMC from donors blood can be directly reprogrammed into iPSC during 21 days period. Patient and healthy donors peripheral blood also can be a source of CD34+ cells subset that can be reprogrammed into iPSC.[304] PBMC derived T cells activated for 5 days with anti-CD3 antibody and IL-2 also can be used for the purpose.[305] In addition, human fibroblasts can be utilized for iPSC creation.[12] The system for such reprogramming is commercially available from ThermoFisher Scientific as CTS™ CytoTune™-iPS 2.1 Sendai Reprogramming Kit, Catalog number: A34546.[306] The relevant video that explains the process of the vector creation entitled "How Does Sendai Virus Reprogram Cells? " is available online. Deriving naive human iPSCs using Sendai virus vectors presents challenges, but these are gradually being overcome.[307]
Airway gene transfer
[edit]SeV vector is one of the most efficient vectors for airway gene transfer. In its natural hosts, like mice, and non-natural hosts, like sheep, SeV-mediated foreign gene expression can be visualized in lungs. This expression is transient: intensive during a few days after the first SeV administration but is returning to baseline, zero values, by day 14. After the second administration, the expression of trans genes is getting reduced by 60% when compared with levels achieved after a first dose.[78]
A replication-defective and persistent Sendai virus can be used as a platform for a durable expression of microRNAs, which were able to inhibit expression of targeted genes.
For vaccine creation
[edit]SeV has several features that are important in a vector for a successful vaccine: the virus does not integrate into the host genome, it does not undergo genetic recombination, it replicates only in the cytoplasm without DNA intermediates or a nuclear phase. SeV, as all other representatives of family Paramyxoviridae, is genetically stable and evolves very slowly. SeV genome can accommodate foreign genes in multiple intergenic positions and the SeV genome is suitable for introducing genes encoding the envelope glycoproteins of pathogenic viruses.[14] For vaccination purpose the virus-based constructs could be delivered in a form of nasal drops, which may be beneficial in inducing a mucosal immune response. This form of vaccination is more immunogenic than intramuscular considering pre-existing anti-SeV antibodies.[309] Sendai virus-based constructs can induce durable, mucosal, B-cell, and T-cell immune responses.[14] The virus genome has high similarity with human parainfluenza virus 1 (HPIV-1) and the two viruses share common antigenic determinants. The study that was published in 2011 demonstrated that SeV neutralizing antibodies (which were formed due to human parainfluenza virus type 1 past infection) can be detected in 92.5% of human subjects worldwide with a median EC50 titer of 60.6 and values ranging from 5.9–11,324.[67] Low anti-SeV antibodies background does not block the ability of SeV-base vaccine to promote antigen-specific T cell immunity.[68]
Wild type, attenuated SeV has been used in clinical trials involving both adults[64] and children[66] to immunize against HPIV-1.The virus administration in the form of nasal drops in doses ranging from 5 × 105 50% embryo infectious dose (EID50) to 5 × 107 induced the production of neutralizing antibodies to the human virus without any measurable side effects. The results of these trials represent an evidence of safety for humans of replication competent Sendai virus administration. SeV antibodies that cross-reactive with HPIV-1 antibodies are present in most people, however, majority of people do not have high titer of these antibodies. The study that was published in 2011 demonstrated that SeV neutralizing antibodies (which were formed due to HPIV-1 past infection) can be detected in 92.5% subjects worldwide with a median EC50 titer of 60.6 and values ranging from 5.9–11,324.[67] Low anti-SeV antibodies background does not block the ability of SeV-base vaccine to promote antigen-specific T cell immunity.[68]
The development of T cell-based AIDS vaccines using Sendai virus vectors is taking place reached phase II clinical trial. Evaluation of the safety and immunogenicity of an intranasally administered replication-competent Sendai Virus–vectored HIV Type 1 gag vaccine demonstrated: induction of potent T-Cell and antibody responses in prime-boost regimens.[18][17]
Sendai virus was also used as a backbone for vaccine against respiratory syncytial virus (HRSV).[13][310] This virus (HRSV), is a major cause of lower respiratory tract infections and hospital visits during infancy and childhood. It was shown that administration of SeV-based RSV vaccine protects cotton rats[311] and African green monkeys from this viral infection.[310] The HRSV phase I clinical trial was completed in adults. It demonstrated high safety of the SeV-based construct that expressed HRSV envelope F glycoprotein.[16]
SeV is currently used in preclinical studies as a backbone vector for vaccine against tuberculosis. Mucosal vaccination with SeV construct generates memory CD8 T cell immunity and promotes protection against Mycobacterium tuberculosis in mice.[15][312][313]
For effective prevention of infections caused by SARS-CoV-2, the ability of the vaccine to stimulate the mucosal immunity of the upper respiratory tract, including the nasal cavity, might be highly important. Such immunity is able to strengthen the antiviral barrier in the upper respiratory tract and provide reliable protection against COVID-19.[314][315] It has been demonstrated that intranasally administered SeV can elicit strong mucosal immunity. Thus, mucosal vaccination with SeV generates robust IgA and IgG antibodies production by nasal-associated lymphoid tissue and by lungs of cotton rats. These antibodies facilitated rapid protection against human parainfluenza virus-type 1.[316]
In China, Fudan University in collaboration with Pharma Co. Ltd. is engaged in development of the vaccine for COVID-19 prevention. SeV serves as a backbone vector in the project [24]. Researchers from the Fudan University have significant experience working with SeV vectors; they created SeV based vaccine for tuberculosis prevention, which is in pre-clinical testing.[317][15][312] There are two Sendai virus strains in China that were described in scientific publications. One of them is BB1 strain,[318] which derived from the Moscow virus strain[147] and has less than 20 non-synomic substitutions compared to Moscow strain. The strain BB1 was given to the researchers of Institute of Viral Disease Control and Prevention, Beijing, China by researchers of Ivanovsky Institute of Virology, Moscow, Russia in the 1960s.[319] Another strain is Tianjin strain, isolated in China in 2008.[319] One of these strains was used for creation of replication deficient SeV85AB construct that is lacking fusion protein (F)[317][15][312] but has inserted sequence encoding immunodominant antigen of Mycobacterium tuberculosis.[320] The safety and immunogenicity of this construct was tested in animal models.[317][15][312] This construct can be easily transformed into the construct that encodes S-protein of SARS-CoV-2. In Russia, State Research Center of Virology and Biotechnology VECTOR is in developing stage of vaccine against COVID-19 using Moscow strain of Sendai virus[147] as a vector backbone.
In Japan researchers have developed two intranasal vaccine candidates against SARS-CoV-2. One design utilizes a modified Sendai virus (SeV) as a vector to deliver the SARS-CoV-2 spike protein's receptor binding domain (RBD) directly to the respiratory tract. In pre-clinical studies, mice received the vaccine intranasally. Mice demonstrated elevated levels of antibodies specific to the SARS-CoV-2 S-RBD (IgM, IgG, IgA) in both their blood serum and bronchoalveolar lavage fluid, lasting up to 12 weeks.[19] Another design uses a similar F-gene lacking SeV vector but different SARS-CoV-2 antigens. Instead of the S-protein, the vaccine design used SARS-CoV-2 nucleocapsid (N), membrane (M), and envelope (E) proteins as immunogens. The study found strong CD8+ T cell responses against these antigens, suggesting intranasal vaccination can trigger immune cells (CD8+ T cells) that target the virus and help control SARS-CoV-2 infection. Vaccination significantly reduced viral load in nasopharyngeal swabs of macaques on day 2 post-challenge, compared to the unvaccinated control group.[321]
Virus biology and properties
[edit]Virion structure
[edit]
Virion structure is well described in a published review.[4] Sendai virus is an enveloped virus: its outer layer is a lipid envelope, which contains glycoprotein hemagglutinin-neurominidase (HN)[322] with two enzymatic activities (hemagglutinating and neuraminidase).[323] Hemagglutinin (H) serves as a cell attachment factor and membrane fusion protein. Neuraminidase (NA) is a sialidase that cleaves and removes sialic acid from the surface of a host cell. This cleavage promotes the fusion of the viral lipid envelope with the cell outer membrane.
In the lipid envelope of the virus located also a fusion protein (F),[324] which is also a glycoprotein that ensures the virus entry into a host cell after viral adsorption. F-protein, as other paramyxoviral fusion proteins, is a trimeric class I viral membrane fusion protein. It is produced in the form of an F0 precursor that must be cleaved by host cell proteases into disulfide-bonded F1 and F2 subunits in order for the trimer to become biologically active.[325] Under the lipid membrane is a matrix protein (M);[326] it forms the inner layer of the virus envelope and stabilizes it structure. The SeV virion also contains the nucleocapsid core, which is composed of the genomic RNA, the nucleocapsid protein (NP),[327] the phosphoproteins (P),[328] which is an essential subunit of the viral of RNA-dependent RNA polymerase (RDRP), and the large protein (L)[329] that is a catalytic subunit of this polymerase. C-protein, which is translated from an alternative reading frame of the P-coding mRNA, is also associated with a viral capsid.[330] It is present in SeV virions at relatively low levels (40 molecules/genome).[331]
Genome
[edit]
Structure
[edit]
The positions of translation initiation sites for products of the alternative reading frame of the P-coding mRNA
The SeV genome is non-segmented, negative-sense RNA, of about 15.384 n. in length, and contains the noncoding 3’ leader and 5’ trailer regions, which are about 50 nucleotides in length.[4][273] As in other respiroviruses from family Paramyxoviridae, in SeV they work as cis-acting elements essential for replication. A 3’ leader sequence acts as a transcriptional promoter. Between these non-coding regions are located six genes, which encode the nucleocapsid (NP) protein, phosphoprotein (P), matrix protein (M), fusion protein (F), hemagglutinin-neuraminidase (HN) and large (L) protein in this order from the 3’ terminus.[4][273] The RNA-dependent RNA polymerase of the SeV consists of the large protein (L) and the phosphoprotein (P). The structural gene sequence of SeV is as follows: 3′-NP-P-M-F-HN-L-5′. Intergenomic regions between these genes are three nucleotides long as in other respiroviruses. Additional proteins, which are frequently called non structural or accessory proteins can be produced from the P gene, using alternative reading frames.[4][332] The Sendai virus P/C mRNA contains five ribosomal initiation sites between positions 81 and 201 from the 5' end. One of these sites initiates in the P open reading frame, whereas four others initiate a nested set of C proteins (C', C, Y1, Y2).[333][332][334] These C proteins are initiated in the + 1 reading frame to that of P at different translation starting sites. Sendai virus uses ribosome shunting to express Y1 and Y2 proteins that initiate at the fourth and fifth start sites on the P/C mRNA (respectively).[334] Three additional SeV proteins are also encoded by P/C mRNA. Two of these proteins V and W are products of RNA editing, at codon 317 of the mRNA - G residues are added co-transcriptionally, (+one G residue for V and +two G for W).[331] The third - X protein is represented by 95 amino acids of the C terminal of the P protein and independently initiated by ribosomes.[335] All these non-structural proteins have several functions, including the organization of viral RNA synthesis and helping the virus to infect rodent cells by escaping host innate immunity (see "The mechanism of viral immunosuppression in natural hosts" section above).[331] It has also been found that C protein facilitates budding of virus-like particles[336] and small amounts of C protein are associated with a viral capsid.[330]
Evolution Stability
[edit]The genomes of non segmented negative-stranded RNA viruses (including paramyxoviruses) have a low rate of homologous recombination and evolve comparatively slowly.[274][275] Multiple reasons for this genomic stability likely exist: (1) the genomes of these viruses are nonsegmented, therefore cannot undergo genetic reassortment, (2) each protein and each amino acid has an important function. Therefore, any new genetic insertion, substitution or deletion would lead to a decrease or total loss of function that would in turn cause the new virus variant to be less viable. (3) Sendai virus belong to viruses that are governed by the “rule of six”. SeV genome as genomes of other paramyxoviruses mainly include six genes, which encode for six major proteins.[276] Low rate of homologous RNA recombination in paramyxoviruses probably results from this unusual genomic requirement for polyhexameric length (6n+0). Natural high genomic stability of SeV is a positive feature for it potential use as a vaccine vector or as an oncolytic agent. For any clinical or industrial applications, it is important that SeV genomic and inserted foreign trans genes would be expressed in a stable way. Paramyxoviruses show relatively little genomic or antigenic change over time. They are known to stably express a wide variety of heterologous antigens at relatively high levels in many species of animals.[277][127][278]
Viral proteins
[edit]| Name and UniProt link | Alias | Function | Category |
|---|---|---|---|
| Nucleocapsid protein | NP | NP protein forms core structure with viral genomic RNA. | structural proteins |
| Phosphoroprotein | P | P-protein is a subunit of the viral of RNA-dependent RNA polymerase. | |
| Matrix protein | M | Matrix protein forms the inner layer of the virus envelope and stabilizes it structure. | |
| Fusion protein | F | Envelope glycoprotein F promotes the fusion of the viral lipid envelope with the cell outer membrane and promotes cell-cell fusion. | |
| Hemagglutinin Neuraminidase | HN | Envelope glycoprotein HN is involved in receptor recognition, sialidase activity, promotes the fusion of the viral lipid envelope with the cell outer membrane, promotes cell-cell fusion. | |
| Large protein | L | L protein represents catalytic subunit of RNA-dependent RNA polymerase.The RNA-dependent RNA polymerase of the virus consists of the large protein (L) and the phosphoprotein (P). | |
| C-protein | C | This protein interacts with IKKα serine / threonine kinase and prevents the phosphorylation of IRF7.[39][40][41] C-protein binds the interferon-alpha/beta receptor subunit 2 (IFNAR2). This binding inhibits IFN-α-stimulated tyrosine phosphorylation of the upstream receptor-associated kinases, TYK2 and JAK1.[45] C-protein suppresses the signal transduction pathways of interferon alpha/beta (IFN-α/β) and IFN-γ by binding to the N-terminal domain of STAT1.[48] C-protein inhibits the production of nitric oxide (NO) by murine macrophages that has cytotoxic activity against viruses.[49][50] C-protein inhibits a pathway that includes a Toll-like receptor (TLR7) and TLR9-induction of IFN-alpha, which is specific for plasmacytoid dendritic cells.[44] C-protein is involved into SeV budding and virions cell exit. C-protein facilitates budding by interacting with AIP1/Alix, which is a host protein that is involved in apoptosis and endosomal membrane trafficking.[337] | non structural |
| C'-protein | C' | apoptosis inhibition, host immunity escape and modulation of virions shape[43][41] | |
| Y1-protein | Y1 | ||
| Y2-protein | Y2 | ||
| V-protein | V | It binds MDA5 and inhibit its activation of the IFN promoter.[53][54] It binds RIG-I and TRIM25. This binding prevents downstream RIG-I signaling to the mitochondrial antiviral signaling protein (MAVS) by disrupting TRIM25 -mediated ubiquitination of RIG-I.[55] V-protein suppresses the production of interleukin-1β, by inhibiting the assembly of the inflammasome NLRP3.[57] | |
| W-protein | W | apoptosis inhibition, host immunity escape and modulation of virions shape[43] | |
| X-protein | X |
Proteolytic cleavage by cellular proteases
[edit]The SeV F protein is a type I membrane glycoprotein that is synthesized as an inactive precursor (F0) that must be activated by proteolytic cleavage at residue arginine-116.[4] After the cleavage F0 precursor yields two disulfide-linked subunits F1 and F2.[298] Paramyxoviruses use different host cell proteases to activate their F-proteins. Sendai virus uses activating proteases that are serine endopeptidases represented by tryptase beta 2-(TPSB2),WikiGenes - Collaborative Publishing (which has aliases such as tryptase II, tryptase Clara, club cells tryptase, mast cells tryptase,[338][339][340][341]) trypsin 1 (PRSS1),[342] mini-plasmin (PLG)[343] and transmembrane serine protease 2 (TMPRSS2).[344] Most likely, blood clotting factor X (F10) is capable to cleave and activate SeV F0.[345][346][347] It is possible that other, not yet identified cellular proteases, can also process the F0 protein of SeV.
SeV cell entry receptors
[edit]
To infect host cells SeV must first bind to cell surface receptors using its hemagglutinin-neuraminidase (HN) protein. The receptor-virus attaching process triggers a conformational change in HN, which allosterically promotes the viral fusion (F) protein to promote virus envelope - cell membrane fusion. The receptor attachment is cooperative with respect to receptor density.[348] SeV cell entry receptors are represented mainly by glycoproteins and glycolipids.[349][350] The table below lists all the molecules that have been shown to function as SeV receptors. Human sialoglycoprotein - cluster of differentiation (CD 235a) is an example of glycoproteins that facilitates SeV cell entry. However, other type of proteins that are not glycoproteins also can assist SeV to penetrate cells. Thus, C-type lectin represented by asialoglycoprotein receptor (ASGP-R), ASGR1[169]) has been shown to be able to function as a SeV cell entry receptor.[165][166][351] Among glycosphingolipids two types of glycans are serving as SeV receptors. The first type is represented by fucosylated glycans and the second one by sialylated glycans.[349] The number, positioning, and chemical linkage of sialic acid-containing receptors can be an important determinant of the strength and efficiency of viral attachment, which can play an important role both in host and tissue tropism.[348]
The expression of molecules that can facilitate SeV cell entry, frequently accelerates carcinogenesis and/or metastasis development. The asialoglycoprotein receptor is highly expressed in liver cancers.[167] The presence of Sialyl-Lewisx antigen (cluster of differentiation 15s (CD15s)), which is a fucosylated glycan, on the outer cell membrane, correlates with invasion potential of malignant cells, tumor recurrence, and overall patient survival for an extremely wide range of cancers.[152][153] Expression of the Vim2 antigen, which is another SeV cell entry receptor represented by fucosylated glycan, is very important for the extravascular infiltration process of acute myeloid leukemia cells.[155] Metastatic cancer cells often are coated with glycolipids that are rich in sialic acids.[154] SeV binds to α2,3-linked sialic acid containing glycolipids.[348][349] For example, GD1a,[156] which is a ganglioside and sialylated glycan (glycolipid), is found in large quantities on the surfaces of breast cancer stem cells.[157] High cell surface expression of another SeV receptor - ganglioside sialosylparagloboside /SPG/ NeuAcα2-3PG.[158] characterizes lymphoid leukemia cells.[159][160] Among other receptors represented by gangliosides GT1b is highly expressed on the outer membranes of brain metastases cells that originate from an extremely broad range of cancer,[161] while GD1a,[156] GT1b[162] and GQ1b[163] can be detected in human gliosarcomas. However, their quantity is not exceeding the quantity in normal frontal cerebral cortex.[164]

A fluorescence microscopy-based assay reveals that the relative number of SeV virions bound to the receptor can be defined as 0.5 for GM3, as 1 for GD1a, and as 2 for Gq1b.[348] The structures of some of these receptors are available for visualization through SugarBindDB - a resource of glycan-mediated host–pathogen interactions.[364] Others are available through KEGG Glycan Database,[365] PubChem compound database,[366] and TOXNET database (toxicology data network) of US National Library of Medicine.[367]

Life cycle
[edit]Because SeV is a negative-strand RNA virus the virus entire life cycle is completed in the cytoplasm using its own RNA polymerase.

Adsorbtion and fusion
[edit]Sendai virus initiates infection process by host cell adsorbtion mediated by the recognition of specific receptor molecules.[350] Hemagglutinin neuraminidase (HN) serves as a virus cell attachment protein that interacts with a specific cell entry receptor. NH has sialidase activity, and it is capable of cleaving sialic acid residues from the cell receptor. This cleavage triggers the fusion process of viral envelope and cell membrane, which promotes by cooperation of NH with the viral fusion protein (F).[368] The SeV F-protein as other Paramyxovirus structural fusion proteins is a trimeric molecule that belongs to class I viral membrane fusion proteins.[325] To perform the fusion function F protein must be proteolytically activated from it precursor inactive form F0.[369] This activation requires F0 cleavage by host serine protease before the virus adsorbtion (see the section “proteolytic cleavage by cellular proteases”). F0 must be cleaved by the host protease into F1 and F2 subunits that remained connected through a disulfide covalent bond. The cleavage site in the F0-protein is located N-terminal to the fusion peptide which has N-terminal Hepta-Repeat 1 (HR1) and C-terminal Hepta-Repeat 2 (HR2) domains. The illustration below shows 5 stages of the fusion of the virus envelope and cellular host membrane.[325] 1) The pre-fusion, F protein (highlighted in red) is protruding from the lipid bilayer of the viral envelope and is in a close proximity to the cellular membrane. 2) Receptor-HN binding, during the SeV host cell attachment process, triggers the release of the fusion peptide from the F-protein. The peptide inserts itself into the host cell membrane. This insertion is accompanied by the transformation of the HR1 domain from a helical structure to an extended helical trimeric coil-coil structure. 3) Transformed HR1 domain attaches viral F-protein to the host-cell membrane. 4) Two lipid bilayers (viral and cellular) fuse with each other. 5) The fusion of the HR2 and HR1 domains of the F-protein promotes the establishment of a stable six-helix bundle structure (6HB). The formation of the 6HB structure leads to the establishment of the pore and the completion of the fusion process. Viral genomic material enters the host cell through this formed pore.[325]
Uncoating
[edit]After a merging of the host membrane and the viral envelope, the SeV according to one model is “uncoating” with diffusion of the viral envelope proteins into the host plasma membrane.[370] According to another model the virus did not release its envelope proteins into the host membrane. The viral and host membranes are fused and a connecting structure is made. This connecting structure serves as a transportation "highway" for the viral ribonucleoprotein (RNP). Thus, RNP travels through the connecting structure to reach the cell interior[370] allowing SeV genetic material to enter the host cell cytoplasm.[368][371]
Cytoplasmic transcription and replication
[edit]Once in the cytoplasm, the SeV genomic RNA is getting involved, as a template, in two different RNA synthetic processes performed by RNA-dependent RNA polymerase, which consists of L and P proteins: (1) transcription to generate mRNAs and (2) replication to produce a positive-sense antigenome RNA that in turn acts as a template for production of progeny negative-strand genomes.[372][373] RNA-dependent RNA polymerase promotes the generation of mRNAs methylated cap structures.[374]
The NP protein is thought to have both structural and functional roles[375] This protein concentration is believed to regulate the switch from RNA transcription to RNA replication. The genomic RNA functions as the template for the viral RNA transcription until the NP protein concentration increases. As the NP protein accumulates, the transition from the transcription to the replication occurs.[376] The NP protein encapsidates the genomic RNA, forming a helical nucleocapsid which is the template for RNA synthesis by the viral RNA polymerase. The protein is a crucial component of the following complexes NP-P (P, phosphoprotein), NP-NP, nucleocapsid-polymerase, and RNA-NP. All these complexes are needed for the viral RNA replication.[375] Research has demonstrated that de novo synthesis of the NP and P proteins is essential for efficient viral transcription. Studies using SeV mutants deficient in NP or P proteins, with genome replication disabled, revealed that the P protein is necessary for viral gene expression. Furthermore, the NP protein plays a crucial role in enhancing primary transcription. Without newly synthesized NP protein, early transcription occurs at minimal levels, while its presence restores normal transcriptional activity. These findings indicate that the synthesis of these proteins facilitates the transition from preliminary to efficient transcription, underscoring their importance in the viral life cycle.[377]
Translation
[edit]Two different sets of proteins are translated from viral mRNAs.[4] The first set is represented by six structural proteins that include nucleocapsid protein (NP), phosphoprotein (P), matrix protein (M), fusion protein (F), neuraminidase (NA) and large protein (L).[4] All these proteins have variable functions and are incorporated into the viral capsid (see the section “virion structure” above). The second set is represented by seven non structural or accessory proteins.[4] These proteins are translated from the polycistronic mRNA of P gene.[333][332][334] This mRNA encodes eight translation products, and P-protein is only one of them. Alternative variants of translation are represented by V,W, C, C’,Y,Y’ and X proteins. The proteins C’, C, Y1, Y2 are products of mRNA alternative reading frame, they collectively referred as C-proteins or C-nested proteins and they share common C-terminal end.[4][378] The X protein also shares the same C-terminal end and its translation also independently initiated by ribosomes.[335] The proteins V and W are products of cotranscriptional mRNA editing. All these non-structural proteins have multiple functions, including the organization of viral RNA synthesis and helping the virus to infect host cells by escaping host innate immunity[331] (see "The mechanism of viral immunosuppression in natural hosts" section above).

Transportation of RNP and viral proteins to cellular membrane
[edit]After translation, SeV nucleocapsids (RNP complex) assemble, and move using microtubules network through intracellular vesicular trafficking pathway.[289] In preparation for the budding process, three viral lipophilic proteins HN, F and M migrate through the secretory pathway to a host cell membrane.[289][379] It is assumed that the interaction of these three proteins with each other is needed for their migration to cellular budding sites.[289] The binding of lipophilic protein complex to the host membrane facilitates the interaction of this three protein complex with the SeV nucleocapsid.[379] It has been shown that for efficient virion production SeV induces the β-cytoplasmic actin remodeling in its host cell.[380]
Syncytium formation and direct cell-to-cell infection transmission
[edit]Two of SeV proteins: HA and F, after their binding directly to a cellular membrane, promote a cell-cell fusion, which leads to a large multinuclear cell formation (syncytium). This formation involves the fusion of infected cells with adjacent target cells and remains an important mechanism of direct cell-to-cell spread of viral components. Thus, a SeV infection in a form of genetic material in partially assembled virions can spread without any exposure to host neutralizing antibodies (see the section "Directed cells fusion (syncytium formation)" for details and references).
Budding
[edit]Sendai virus, as all other envelope viruses, uses host cellular membrane lipid bilayer for viral capsid membrane formation. Binding to a host cell membrane of viral proteins (M, HN and F) promotes their interaction with RNP complex, which is composed of the viral genomic RNA bound to SeV proteins (NP, P and L).[379] Thus, all viral structural components, including viral glycoproteins and genomic RNP complex, are getting assembled together. After such assembling the infectious viral particles are budding out from individually or collectively infected cells (syncitia). It has been suggested that recirculating endosomes are involved in viral RNP complex translocation.[289] C-protein facilitates budding by interacting with AIP1/Alix, which is a host protein that is involved in apoptosis and endosomal membrane trafficking.[337] The infectious virus particles usually released by 24 hours post infection (hpi), and peak titers appeared between 48 and 72 hpi.[292]
Persistent infection
[edit]Sendai virus can establish persistent infection in its host cells. Multiple rounds of virus subculturing result in a creation of new virus variants with high ability to establish persistent infection. These SeV variants develop certain genotypic changes.[381] Specific amino acid substitutions accumulated in the M protein and the L protein were show to be associated with persistent infection in mouse connective tissue cells (L-929) and hamster kidney fibroblasts (BHK-21).[381] It has been shown that 4-5 point mutations might distinguish Sendai variants capable of persistent infections from those that are incapable.The most common single nucleotide mutations are found in the leader sequence (position 16) and in the following genes: the M gene (position 850), the F gene (position 782), and the L gene (positions 832 and 1743).[382]
The persistent infection can also be established instantly in interferon regulatory factor 3 (IRF-3)-knockdown cells. IRF-3 is a key proapoptotic protein that after activation by SeV triggers apoptosis. IRF-3-knockdown cells express viral protein and produces low levels of infectious virions.[383][384] IRF-3 controls the fate of the SeV-infected cells by triggering apoptosis and preventing persistence establishment; therefore its knock down allows persistence to occur.[116] It was also reported that during SeV infection replication defective viral genomes (DVG) are forming[385] and selectively protect a subpopulation of host cells from death, therefore promoting the establishment of persistent infections.[386][387] The possibility of establishing a chronic viral infection was further demonstrated in Sendai virus-infected ovine fibroblasts.[388] In nature enzootic disease patterns suggest that the virus can be latent and can be cleared over the course of a year.[389]
Directed cells fusion (syncytium formation)
[edit]One recognized feature of the Sendai virus, shared with members of its genus, is the ability to induce syncytia formation in vivo and in vitro in eukaryotic cell cultures.[390] The formation of syncytium helps the virus to avoid neutralizing antibodies of the host organism during the spread of infection. The mechanism for this process is fairly well understood and is very similar to the fusion process employed by the virion to facilitate cellular entry. The activities of the receptor binding hemagglutinin-neuraminidase protein is solely responsible for inducing close interaction between the virus envelope and the cellular membrane.
However, it is the F protein (one of many membrane fusion proteins) that, when triggered by local dehydration[391] and a conformational change in the bound HN protein,[392] actively inserts into the cellular membrane, which causes the envelope and the membrane to merge, followed shortly by virion entry. When the HN and F protein are manufactured by the cell and expressed on the surface, the same process may occur between adjacent cells, causing extensive membrane fusion and resulting in the formation of a syncytium.[393]
Using the model of cellular hepatocarcinoma (Hep G2), it has been shown that Sendai virus recruits the cellular protein villin for cell fusion and syncytia formation. The villin-actin interaction regulates the fusion of the viral envelope and the cell membrane. Thus, villin is a host cell cofactor that regulates the fusion process.[394] Its down-regulation with siRNA inhibits SeV infection of Hep G2 cells.[394]
The cell fusion property of SeV was utilized by Köhler and Milstein, who published an article in 1975 outlining a revolutionary method of manufacturing monoclonal antibodies. In need of a reliable method to produce large quantities of a specific antibody, the two merged a monoclonal B cell, exposed to a chosen antigen, and a myeloma tumor cell to produce hybridomas, capable of being grown indefinitely and of producing significant amounts of an antibody specifically targeting the chosen antigen. Though more efficient methods of creating such hybrids have since been found, Köhler and Milstein first used Sendai virus to create their revolutionary cells.[10]
Sensitive cell lines, primary cultures and virus strains
[edit]
Cell lines
[edit]
Scientific studies show that the following cell lines are susceptible to SeV infection to varying degrees.
Some of these cells (for example, LLC MK2, 4647 and HEK 293) do not express a protease that processes fusion protein F0 of Sendai virus; therefore, they produce non-infectious virions.[400]
Type 1 IFN inhibits the SeV production in normal human respiratory cells,[79] but fails of doing it in human cells that originates from variable malignancies such as U937,[225] Namalwa,[225] and A549.[225]
Variable cell cultures obtained from tumors have different sensitivity to SeV, and can also produce the virus in different quantities.[396] There are multiple factors that are responsible for this variability. For example, an inverse correlation was observed between cells sensitivity to SeV infection and constitutive mRNA expression levels of TLR 3 and TLR 7 in primary cultures of prostate cancer.[402] Thus, defective TLR-activated IFN signaling is one of these factors.

Virus adaptation to grow in cell cultures
[edit]SeV strain variants adapted for growth in different cells have different properties. One study shows that the SeV variant adapted for growth in LLC-MK2 cells and the SeV variant adapted for growth in embryonated eggs differ by two amino acids in the HN protein. This difference results in different neuraminidase conformations around the receptor binding site and variations in neuraminidase activity between the two viral variants.[403] Another research study shows that SeV variants, adapted to grow in cell culture 4647 (African green monkey kidney cells) and in HEK 293 (human embryonic kidney cells) instead of embryonated chicken eggs, also acquire mutations in HN gene and both SeV variants lost their oncolytic activity.[400][404]

Primary cultures
[edit]Ovine blood-derived and alveolar macrophages can be infected with SeV ex vivo. Experiments with a virus construct with an inserted green fluorescent protein (SeV-GFP) showed that infection reaches 100% of cells in 48 hours. Primary cell cultures of ovine skin fibroblasts can also be infected and also achieve 100% GFP positivity. In fibroblasts, an intracellular virus-associated GFP expression was stable at least for more than a dozen passages in cell culture. However, an infectious virus was not produced in these ovine cells. This fact was demonstrated by the transfer of supernatants from SeV-infected cells into fresh cultures.[388] In addition, human skin fibroblasts can be infected with Sendai virus.[11][12] SeV can replicate to high titers in human monocyte-derived DCs.[105][270]
Persistent infection
[edit]Most often, SeV infection initiates an apoptotic program in the host cells, which leads to the death of target cells without interrupting the life cycle of the virus. However, paramyxoviruses, including SeV, can cause persistent infection in primary cell cultures that does not kill cells or turn off cellular RNA transcription and translation. It has been demonstrated that mouse connective tissues cells (L-929) and hamster kidney fibroblasts (BHK-21) can become infected with Sendai virus and the infection can be persistent.[381] The possibility of establishing a persistent viral infection was demonstrated in SeV-infected ovine fibroblasts.[388]
Strains
[edit]History
[edit]All Sendai virus strains belong to the same serotype. The origin of many strains of SeV was described in 1978.[72] Some strains such as Ohita[403] and Hamamatsu[405] were described later. Ohita and Hamanatsu strains were isolated from separate epidemics in laboratory mice.[406][407] According to the personal memory of Alisa G. Bukrinskaya, who has co-authored numerous publications related to SeV along with Prof. Viktor M. Zhdanov, starting in 1961,[408] the Moscow strain of SeV[147] was obtained by Prof. Viktor M. Zhdanov of the Ivanovsky Institute of Virology from Japan in the late 1950s or early 1960s,[408] It is reported[319] that the BB1 strain[318] derived from the Moscow virus strain.[147] The strain BB1 was given to the researchers of Institute of Viral Disease Control and Prevention, Beijing, China by researchers of Ivanovsky Institute of Virology, Moscow, Russia in the 1960s.[319]
Virulence
[edit]A field SeV isolate, that is attenuated through egg-passages, is less virulent for mouse respiratory cells.[409] Therefore, the strains that were isolated from animals a few decades ago and went through multiple passages in eggs are less virulent for mice in comparison with the strains that are fresh field isolates.
Defective interfering genomes or particles
[edit]Defective interfering (DI) genomes or defective viral genomes (DVGs) or defective interfering particles (DIPs) are replication defective viral RNA products generated during viral infections by many types of viruses, including SeV.[410][385][387] It has been experimentally established that DI genomes can be readily produced by viral infection at high multiplicity.[411][412] A single amino acid substitution in a nucleoprotein (NP) causes an increased production rate of DI genomes in the SeV Cantell strain, which is known for its particularly strong induction of interferon beta (IFN-β) during viral infection.[413] It has been shown that DI are responsible for this strong IFN-β induction.[414] Other genomic change such as loss of the Sendai virus C-protein has also been demonstrated to cause accumulation of DI genomes.[415]
Strains origin and sequence ID
[edit]| Strain name | Origin | Sequence ID |
| Z (Sendai/52, or VR-105, or Fushimi) | derivative of murine isolate of 50s (Japan) | AB855655.1 |
| Cantell (VR-907) | derivative of the same isolate as above | AB855654.1 |
| Enders | derivative of the same isolate as above | * |
| Nagoya | derivative of the same isolate as above | AB275417.1
AB195968.1 |
| Moscow | derivative of murine isolate of 50-60s (Japan or Russia) | KP717417.1 |
| BB1 | derivative of the same murine isolate of 50-60s as above (Japan or Russia) | DQ219803.1 |
| Ohita | murine isolate of 70-90s (Japan) | NC_001552.1 |
| Hamamatsu | independent from Ohita, murine isolate of 70-90s (Japan) | AB039658 |
*The sequence of Enders strain is available from the US patent Modified Sendai virus vaccine and imaging vector
Strains sequence similarity
[edit]| Strain name | Z | Cantell | Enders | Nagoya | Moscow | BB1 | Ohita | Hamamatsu |
| Sendai Virus | Megablast homology for SeV complete genome (%) | |||||||
| Z | 100 | |||||||
| Cantell | 99.3 | 100 | ||||||
| Enders | 99.4 | 99.2 | 100 | |||||
| Nagoya | 98.9 | 100 | ||||||
| Moscow | 88.1 | 88.6 | 87.9 | 100 | ||||
| BB1 | 88.1 | 99.9 | 100 | |||||
| Ohita | 88.9 | 91.2 | 100 | |||||
| Hamamatsu | 91.7 | 91.7 | 99.2 | 100 | ||||
| Human parainfluenza Virus 1 | Discontiguous megablast for complete viral genomes (%) seq. ID AF457102.1 | |||||||
| HPV1 (strain Washington/1964) | 75.2 | 73.9 | 74.5 | 74.6 | ||||
| Porcine parainfluenza virus 1 | Discontiguous megablast for complete viral genomes (%) seq.ID NC_025402.1 | |||||||
| PPV1 (strain S206N) | 71.15 | 75.1 | 70.5 | 71 | ||||
Virus preparation and titration
[edit]Sendai virus can be produced using specific pathogen-free (SPF) embryonated chicken eggs.[416] Sendai virus, adapted to grow in cell culture instead of chicken eggs, loses its oncolytic activity.[400][404]
The Sendai virus titer can be evaluated by serial end point 10x dilution assay of the virus-containing material in embryonated chicken eggs. This assay evaluates the final dilution that may cause a viral infection in 50% of inoculated eggs. This EID50 assay is used to quantify titer for many viruses that can be grown in eggs.[417][418] The measurement of virus titer obtained from this assay is expressed as an embryonic infectious dose 50% (EID50). The SeV titer can also be assessed by using plaque assay in LLC-MK2 cells[419] and by serial end point 2x dilution hemagglutination assay (HA).[420] However, the HA test is less reliable than the EID50 or PFU tests because it does not always indicate the presence of a viable virus in a sample. The dead virus might demonstrate high HA titers.
References
[edit]- ^ Walker P (15 June 2015). "Implementation of taxon-wide non-Latinized binomial species names in the family Rhabdoviridae" (PDF). International Committee on Taxonomy of Viruses (ICTV). p. 7. Retrieved 6 February 2019.
- ^ Samal SK (2008). "Paramyxoviruses of Animals". Encyclopedia of Virology. Elsevier. pp. 40–47. doi:10.1016/b978-012374410-4.00460-x. ISBN 9780123744104. S2CID 81060576.
- ^ "Paramyxoviridae". UniProt.
- ^ a b c d e f g h i j k l m n Faísca P, Desmecht D (February 2007). "Sendai virus, the mouse parainfluenza type 1: a longstanding pathogen that remains up-to-date". Research in Veterinary Science. 82 (1): 115–125. doi:10.1016/j.rvsc.2006.03.009. PMID 16759680.
- ^ "Taxonomy - Respirovirus". UniProt.
- ^ "Respirovirus". ViralZone.
- ^ a b Saga K, Kaneda Y (2015). "Oncolytic Sendai virus-based virotherapy for cancer: recent advances". Oncolytic Virotherapy. 4: 141–7. doi:10.2147/OV.S66419. PMC 4918391. PMID 27512677.
- ^ a b c d e f Matveeva OV, Kochneva GV, Netesov SV, Onikienko SB, Chumakov PM (April 2015). "Mechanisms of Oncolysis by Paramyxovirus Sendai". Acta Naturae. 7 (2): 6–16. doi:10.32607/20758251-2015-7-2-6-16. PMC 4463408. PMID 26085940.
 Material was copied from this source, which is available under a Creative Commons Attribution License.
Material was copied from this source, which is available under a Creative Commons Attribution License.
- ^ a b c Ilyinskaya GV, Mukhina EV, Soboleva AV, Matveeva OV, Chumakov PM (2018). "Oncolytic Sendai Virus Therapy of Canine Mast Cell Tumors (A Pilot Study)". Frontiers in Veterinary Science. 5: 116. doi:10.3389/fvets.2018.00116. PMC 5995045. PMID 29915788.
- ^ a b Köhler G, Milstein C (August 1975). "Continuous cultures of fused cells secreting antibody of predefined specificity". Nature. 256 (5517): 495–497. Bibcode:1975Natur.256..495K. doi:10.1038/256495a0. PMID 1172191. S2CID 4161444.
- ^ a b c d e Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M (2009). "Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome". Proceedings of the Japan Academy. Series B, Physical and Biological Sciences. 85 (8): 348–62. Bibcode:2009PJAB...85..348F. doi:10.2183/pjab.85.348. PMC 3621571. PMID 19838014.
- ^ a b c d e f Ban H, Nishishita N, Fusaki N, Tabata T, Saeki K, Shikamura M, et al. (August 2011). "Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors". Proceedings of the National Academy of Sciences of the United States of America. 108 (34): 14234–9. Bibcode:2011PNAS..10814234B. doi:10.1073/pnas.1103509108. PMC 3161531. PMID 21821793.
- ^ a b c Russell CJ, Hurwitz JL (2015-12-09). "Sendai virus as a backbone for vaccines against RSV and other human paramyxoviruses". Expert Review of Vaccines. 15 (2): 189–200. doi:10.1586/14760584.2016.1114418. PMC 4957581. PMID 26648515.
- ^ a b c d e f g Russell CJ, Hurwitz JL (May 2021). "Sendai Virus-Vectored Vaccines That Express Envelope Glycoproteins of Respiratory Viruses". Viruses. 13 (6): 1023. doi:10.3390/v13061023. PMC 8230104. PMID 34072332.
- ^ a b c d e Hu Z, Jiang W, Gu L, Qiao D, Shu T, Lowrie DB, et al. (December 2019). "Heterologous prime-boost vaccination against tuberculosis with recombinant Sendai virus and DNA vaccines". Journal of Molecular Medicine. 97 (12): 1685–1694. doi:10.1007/s00109-019-01844-3. PMID 31786669. S2CID 208359634.
- ^ a b c Scaggs Huang F, Bernstein DI, Slobod KS, Portner A, Takimoto T, Russell CJ, et al. (February 2021). "Safety and immunogenicity of an intranasal sendai virus-based vaccine for human parainfluenza virus type I and respiratory syncytial virus (SeVRSV) in adults". Human Vaccines & Immunotherapeutics. 17 (2): 554–559. doi:10.1080/21645515.2020.1779517. PMC 7899675. PMID 32750273.
- ^ a b c Seki S, Matano T (2016). "Development of a Sendai virus vector-based AIDS vaccine inducing T cell responses". Expert Review of Vaccines. 15 (1): 119–127. doi:10.1586/14760584.2016.1105747. PMID 26512881. S2CID 27197590.
- ^ a b c Nyombayire J, Anzala O, Gazzard B, Karita E, Bergin P, Hayes P, et al. (January 2017). "First-in-Human Evaluation of the Safety and Immunogenicity of an Intranasally Administered Replication-Competent Sendai Virus-Vectored HIV Type 1 Gag Vaccine: Induction of Potent T-Cell or Antibody Responses in Prime-Boost Regimens". The Journal of Infectious Diseases. 215 (1): 95–104. doi:10.1093/infdis/jiw500. PMC 5225252. PMID 28077588.
- ^ a b Morimoto S, Saeki K, Takeshita M, Hirano K, Shirakawa M, Yamada Y, et al. (January 2023). "Intranasal Sendai virus-based SARS-CoV-2 vaccine using a mouse model". Genes to Cells. 28 (1): 29–41. doi:10.1111/gtc.12992. PMID 36401755. S2CID 253671438.
- ^ Cassano A, Rasmussen S, Wolf FR (January 2012). "Viral diseases". In Suckow MA, Stevens KA, Wilson RP (eds.). The laboratory rabbit, guinea pig, hamster, and other rodents. Academic Press. pp. 821–837. ISBN 978-0-12-380920-9. American College of Laboratory Animal Medicine.
- ^ MacLachlan NJ, Dubovi EJ, eds. (2017). "Chapter 17 - Paramyxoviridae and Pneumoviridae". Fenner's Veterinary Virology (Fifth ed.). Academic Press. pp. 327–356. doi:10.1016/B978-0-12-800946-8.00017-9. ISBN 9780128009468. S2CID 214757272.
- ^ Flecknell PA, Parry R, Needham JR, Ridley RM, Baker HF, Bowes P (April 1983). "Respiratory disease associated with parainfluenza Type I (Sendai) virus in a colony of marmosets (Callithrix jacchus)". Laboratory Animals. 17 (2): 111–113. doi:10.1258/002367783780959448. PMID 6306336. S2CID 7413539.
- ^ Suckow MA, Stevens KA, Wilson RP (23 January 2012). The Laboratory Rabbit, Guinea Pig, Hamster, and Other Rodents. Elsevier. ISBN 978-0-12-380920-9.
- ^ Nicklas W, Bleich A, Mähler M (2012-01-01). "Chapter 3.2 - Viral Infections of Laboratory Mice". In Hedrich HJ (ed.). The Laboratory Mouse (Second ed.). Academic Press. pp. 427–480. doi:10.1016/B978-0-12-382008-2.00019-2. ISBN 9780123820082. PMC 7150319.
- ^ a b c López CB, Yount JS, Hermesh T, Moran TM (May 2006). "Sendai virus infection induces efficient adaptive immunity independently of type I interferons". Journal of Virology. 80 (9): 4538–4545. doi:10.1128/JVI.80.9.4538-4545.2006. PMC 1472017. PMID 16611914.
- ^ "Sendai Virus". Diseases of Research Animals.
- ^ Faisca P, Anh DB, Desmecht DJ (November 2005). "Sendai virus-induced alterations in lung structure/function correlate with viral loads and reveal a wide resistance/susceptibility spectrum among mouse strains". American Journal of Physiology. Lung Cellular and Molecular Physiology. 289 (5): L777–L787. doi:10.1152/ajplung.00240.2005. PMID 16006482.
- ^ a b c d e f g h Burke CW, Mason JN, Surman SL, Jones BG, Dalloneau E, Hurwitz JL, et al. (July 2011). "Illumination of parainfluenza virus infection and transmission in living animals reveals a tissue-specific dichotomy". PLOS Pathogens. 7 (7): e1002134. doi:10.1371/journal.ppat.1002134. PMC 3131265. PMID 21750677.
- ^ Parker JC, Whiteman MD, Richter CB (January 1978). "Susceptibility of inbred and outbred mouse strains to Sendai virus and prevalence of infection in laboratory rodents". Infection and Immunity. 19 (1): 123–30. doi:10.1128/IAI.19.1.123-130.1978. PMC 414057. PMID 203530.
- ^ a b Brownstein DG, Winkler S (April 1986). "Genetic resistance to lethal Sendai virus pneumonia: virus replication and interferon production in C57BL/6J and DBA/2J mice". Laboratory Animal Science. 36 (2): 126–9. PMID 2422437.
- ^ Simon AY, Moritoh K, Torigoe D, Asano A, Sasaki N, Agui T (December 2009). "Multigenic control of resistance to Sendai virus infection in mice". Infection, Genetics and Evolution. 9 (6): 1253–9. Bibcode:2009InfGE...9.1253S. doi:10.1016/j.meegid.2009.08.011. hdl:2115/42554. PMID 19733691.
- ^ Breider MA, Adams LG, Womack JE (December 1987). "Influence of interferon in natural resistance of mice to Sendai virus pneumonia". American Journal of Veterinary Research. 48 (12): 1746–50. PMID 2449103.
- ^ Sangster M, Smith FS, Coleclough C, Hurwitz JL (September 1995). "Human parainfluenza virus type 1 immunization of infant mice protects from subsequent Sendai virus infection". Virology. 212 (1): 13–9. doi:10.1006/viro.1995.1448. PMID 7676623.
- ^ Stone AE, Giguere S, Castleman WL (November 2003). "IL-12 reduces the severity of Sendai virus-induced bronchiolar inflammation and remodeling". Cytokine. 24 (3): 103–13. doi:10.1016/j.cyto.2003.07.005. PMID 14581004.
- ^ a b c "Sendai Virus (SV)". Rat Guide.
- ^ Kraft V, Meyer B (June 1986). "Diagnosis of murine infections in relation to test methods employed". Laboratory Animal Science. 36 (3): 271–6. PMID 3014210.
- ^ Lock LF (2007). "Mouse and Human Pluripotent Stem Cells". In Fox JG, Barthold S, Davisson M, Newcomer CE, Quimby FW, Smith A (eds.). The Mouse in Biomedical Research (2nd ed.). Amsterdam: Academic Press. pp. 281–309. doi:10.1016/B978-012369454-6/50039-X.
- ^ Eaton GJ, Lerro A, Custer RP, Crane AR (August 1982). "Eradication of Sendai pneumonitis from a conventional mouse colony". Laboratory Animal Science. 32 (4): 384–6. PMID 6292576.
- ^ a b c Kiyotani K, Sakaguchi T, Kato A, Nagai Y, Yoshida T (March 2007). "Paramyxovirus Sendai virus V protein counteracts innate virus clearance through IRF-3 activation, but not via interferon, in mice". Virology. 359 (1): 82–91. doi:10.1016/j.virol.2006.08.053. PMID 17027894.
- ^ a b c Irie T, Nagata N, Igarashi T, Okamoto I, Sakaguchi T (May 2010). "Conserved charged amino acids within Sendai virus C protein play multiple roles in the evasion of innate immune responses". PLOS ONE. 5 (5): e10719. Bibcode:2010PLoSO...510719I. doi:10.1371/journal.pone.0010719. PMC 2873429. PMID 20502666.
- ^ a b c d Kato A, Ohnishi Y, Kohase M, Saito S, Tashiro M, Nagai Y (April 2001). "Y2, the smallest of the Sendai virus C proteins, is fully capable of both counteracting the antiviral action of interferons and inhibiting viral RNA synthesis". Journal of Virology. 75 (8): 3802–10. doi:10.1128/JVI.75.8.3802-3810.2001. PMC 114871. PMID 11264369.
- ^ Popli S, Chakravarty S, Fan S, Glanz A, Aras S, Nagy LE, et al. (September 2022). "IRF3 inhibits nuclear translocation of NF-κB to prevent viral inflammation". Proceedings of the National Academy of Sciences of the United States of America. 119 (37): e2121385119. Bibcode:2022PNAS..11921385P. doi:10.1073/pnas.2121385119. PMC 9478676. PMID 36067309.
- ^ a b c Koyama AH, Irie H, Kato A, Nagai Y, Adachi A (April 2003). "Virus multiplication and induction of apoptosis by Sendai virus: role of the C proteins". Microbes and Infection. 5 (5): 373–8. doi:10.1016/S1286-4579(03)00043-1. PMID 12737992.
- ^ a b c Yamaguchi M, Kitagawa Y, Zhou M, Itoh M, Gotoh B (January 2014). "An anti-interferon activity shared by paramyxovirus C proteins: inhibition of Toll-like receptor 7/9-dependent alpha interferon induction". FEBS Letters. 588 (1): 28–34. Bibcode:2014FEBSL.588...28Y. doi:10.1016/j.febslet.2013.11.015. PMID 24269682. S2CID 24831300.
- ^ a b Kitagawa Y, Yamaguchi M, Kohno M, Sakai M, Itoh M, Gotoh B (2020). "Respirovirus C protein inhibits activation of type I interferon receptor-associated kinases to block JAK-STAT signaling". FEBS Letters. 594 (5): 864–877. doi:10.1002/1873-3468.13670. PMID 31705658. S2CID 207944272.
- ^ a b c Irie T, Yoshida A, Sakaguchi T (2013-08-09). "Clustered basic amino acids of the small sendai virus C protein Y1 are critical to its RAN GTPase-mediated nuclear localization". PLOS ONE. 8 (8): e73740. Bibcode:2013PLoSO...873740I. doi:10.1371/journal.pone.0073740. PMC 3739745. PMID 23951363.
- ^ Oda K, Matoba Y, Irie T, Kawabata R, Fukushi M, Sugiyama M, et al. (November 2015). "Structural Basis of the Inhibition of STAT1 Activity by Sendai Virus C Protein". Journal of Virology. 89 (22): 11487–99. doi:10.1128/JVI.01887-15. PMC 4645678. PMID 26339056.
- ^ a b Oda K, Oda T, Matoba Y, Sato M, Irie T, Sakaguchi T (December 2017). "Structural analysis of the STAT1:STAT2 heterodimer revealed the mechanism of Sendai virus C protein-mediated blockade of type 1 interferon signaling". The Journal of Biological Chemistry. 292 (48): 19752–19766. doi:10.1074/jbc.m117.786285. PMC 5712616. PMID 28978648.
- ^ a b Odkhuu E, Komatsu T, Naiki Y, Koide N, Yokochi T (November 2014). "Sendai virus C protein inhibits lipopolysaccharide-induced nitric oxide production through impairing interferon-β signaling". International Immunopharmacology. 23 (1): 267–72. doi:10.1016/j.intimp.2014.09.012. PMID 25242386.
- ^ a b Odkhuu E, Komatsu T, Koide N, Naiki Y, Takeuchi K, Tanaka Y, et al. (October 2018). "Sendai virus C protein limits NO production in infected RAW264.7 macrophages". Innate Immunity. 24 (7): 430–438. doi:10.1177/1753425918796619. PMC 6830875. PMID 30189760.
- ^ MacMicking J, Xie QW, Nathan C (1997). "Nitric oxide and macrophage function". Annual Review of Immunology. 15 (1): 323–50. doi:10.1146/annurev.immunol.15.1.323. PMID 9143691.
- ^ a b c Takeuchi K, Komatsu T, Kitagawa Y, Sada K, Gotoh B (October 2008). "Sendai virus C protein plays a role in restricting PKR activation by limiting the generation of intracellular double-stranded RNA". Journal of Virology. 82 (20): 10102–10. doi:10.1128/JVI.00599-08. PMC 2566265. PMID 18684815.
- ^ a b Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, et al. (December 2004). "The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter". Proceedings of the National Academy of Sciences of the United States of America. 101 (49): 17264–9. Bibcode:2004PNAS..10117264A. doi:10.1073/pnas.0407639101. PMC 535396. PMID 15563593.
- ^ a b Childs K, Stock N, Ross C, Andrejeva J, Hilton L, Skinner M, et al. (March 2007). "mda-5, but not RIG-I, is a common target for paramyxovirus V proteins". Virology. 359 (1): 190–200. doi:10.1016/j.virol.2006.09.023. PMID 17049367.
- ^ a b Sánchez-Aparicio MT, Feinman LJ, García-Sastre A, Shaw ML (March 2018). "Paramyxovirus V Proteins Interact with the RIG-I/TRIM25 Regulatory Complex and Inhibit RIG-I Signaling". Journal of Virology. 92 (6). doi:10.1128/JVI.01960-17. PMC 5827389. PMID 29321315.
- ^ Morita N, Tanaka Y, Odkhuu E, Naiki Y, Komatsu T, Koide N (February 2020). "Sendai virus V protein decreases nitric oxide production by inhibiting RIG-I signaling in infected RAW264.7 macrophages". Microbes and Infection. 22 (8): 322–330. doi:10.1016/j.micinf.2020.01.005. PMID 32032681. S2CID 211064429.
- ^ a b Komatsu T, Tanaka Y, Kitagawa Y, Koide N, Naiki Y, Morita N, et al. (October 2018). "Sendai Virus V Protein Inhibits the Secretion of Interleukin-1β by Preventing NLRP3 Inflammasome Assembly". Journal of Virology. 92 (19): e00842–18. doi:10.1128/JVI.00842-18. PMC 6146803. PMID 30021903.
- ^ Rochat S, Komada H, Kolakofsky D (July 1992). "Loss of V protein expression in human parainfluenza virus type 1 is not a recent event". Virus Research. 24 (2): 137–44. doi:10.1016/0168-1702(92)90002-q. PMID 1326826.
- ^ Odkhuu E, Komatsu T, Koide N, Naiki Y, Takeuchi K, Tanaka Y, et al. (October 2018). "Sendai virus C protein limits NO production in infected RAW264.7 macrophages". Innate Immunity. 24 (7): 430–438. doi:10.1177/1753425918796619. PMC 6830875. PMID 30189760.
- ^ a b Qiao D, Janke BH, Elankumaran S (August 2009). "Molecular characterization of glycoprotein genes and phylogenetic analysis of two swine paramyxoviruses isolated from United States". Virus Genes. 39 (1): 53–65. doi:10.1007/s11262-009-0353-2. PMID 19337823. S2CID 7100230.
- ^ Qiao D, Janke BH, Elankumaran S (January 2010). "Complete genome sequence and pathogenicity of two swine parainfluenzavirus 3 isolates from pigs in the United States". Journal of Virology. 84 (2): 686–94. doi:10.1128/JVI.00847-09. PMC 2798373. PMID 19906928.
- ^ a b c d Lau SK, Woo PC, Wu Y, Wong AY, Wong BH, Lau CC, et al. (October 2013). "Identification and characterization of a novel paramyxovirus, porcine parainfluenza virus 1, from deceased pigs". The Journal of General Virology. 94 (Pt 10): 2184–90. doi:10.1099/vir.0.052985-0. PMID 23918408.
- ^ a b c Palinski RM, Chen Z, Henningson JN, Lang Y, Rowland RR, Fang Y, et al. (February 2016). "Widespread detection and characterization of porcine parainfluenza virus 1 in pigs in the USA". The Journal of General Virology. 97 (2): 281–286. doi:10.1099/jgv.0.000343. PMID 26581410.
- ^ a b c d e Slobod KS, Shenep JL, Luján-Zilbermann J, Allison K, Brown B, Scroggs RA, et al. (August 2004). "Safety and immunogenicity of intranasal murine parainfluenza virus type 1 (Sendai virus) in healthy human adults". Vaccine. 22 (23–24): 3182–6. doi:10.1016/j.vaccine.2004.01.053. PMID 15297072.
- ^ a b Skiadopoulos MH, Surman SR, Riggs JM, Elkins WR, St Claire M, Nishio M, et al. (May 2002). "Sendai virus, a murine parainfluenza virus type 1, replicates to a level similar to human PIV1 in the upper and lower respiratory tract of African green monkeys and chimpanzees". Virology. 297 (1): 153–60. doi:10.1006/viro.2002.1416. PMID 12083845.
- ^ a b c Adderson E, Branum K, Sealy RE, Jones BG, Surman SL, Penkert R, et al. (March 2015). "Safety and immunogenicity of an intranasal Sendai virus-based human parainfluenza virus type 1 vaccine in 3- to 6-year-old children". Clinical and Vaccine Immunology. 22 (3): 298–303. doi:10.1128/CVI.00618-14. PMC 4340902. PMID 25552633.
- ^ a b c d Hara H, Hara H, Hironaka T, Inoue M, Iida A, Shu T, et al. (June 2011). "Prevalence of specific neutralizing antibodies against Sendai virus in populations from different geographic areas: implications for AIDS vaccine development using Sendai virus vectors". Human Vaccines. 7 (6): 639–45. doi:10.4161/hv.7.6.15408. PMID 21508675. S2CID 24481304.
- ^ a b c d Moriya C, Horiba S, Inoue M, Iida A, Hara H, Shu T, et al. (July 2008). "Antigen-specific T-cell induction by vaccination with a recombinant Sendai virus vector even in the presence of vector-specific neutralizing antibodies in rhesus macaques". Biochemical and Biophysical Research Communications. 371 (4): 850–4. doi:10.1016/j.bbrc.2008.04.156. PMID 18466766.
- ^ Kuroya M, Ishida N (August 1953). "Newborn virus pneumonitis (type Sendai). II. The isolation of a new virus possessing hemagglutinin activity". Yokohama Medical Bulletin. 4 (4): 217–33. PMID 13137076.
- ^ Kuroya M, Ishida N, Shiratori T (June 1953). "Newborn virus pneumonitis (type Sendai). II. The isolation of a new virus". The Tohoku Journal of Experimental Medicine. 58 (1): 62. doi:10.1620/tjem.58.62. PMID 13102529.
- ^ Fukumi H, Nishikawa F, Kitayama T (August 1954). "A pneumotropic virus from mice causing hemagglutination". Japanese Journal of Medical Science & Biology. 7 (4): 345–63. doi:10.7883/yoken1952.7.345. PMID 13232830.
- ^ a b c d e Ishida N, Homma M (1978). "Sendai virus". Advances in Virus Research. 23: 349–83. doi:10.1016/S0065-3527(08)60103-7. ISBN 9780120398232. PMID 219669.
- ^ a b "Sendai virus | infectious agent". Encyclopedia Britannica. Retrieved 2019-08-26.
- ^ "Sendai virus (ATCC VR-105)". American Type Culture Collection (ATCC).
- ^ "Sendai virus". Swine Health Information Center. Iowa State University, Center for Food Security and Public Health, College of Veterinary Medicine. September 2015.
- ^ "Sendai Virus Fact Sheet". Stanford Environmental Health & Safety.
- ^ "Recombinant Sendai Viral Vectors" (PDF). Occupational and Environmental Health and Safety. University of Utah.
- ^ a b Griesenbach U, McLachlan G, Owaki T, Somerton L, Shu T, Baker A, et al. (February 2011). "Validation of recombinant Sendai virus in a non-natural host model". Gene Therapy. 18 (2): 182–8. doi:10.1038/gt.2010.131. PMID 20962870. S2CID 23293412.
- ^ a b c d Bousse T, Chambers RL, Scroggs RA, Portner A, Takimoto T (October 2006). "Human parainfluenza virus type 1 but not Sendai virus replicates in human respiratory cells despite IFN treatment". Virus Research. 121 (1): 23–32. doi:10.1016/j.virusres.2006.03.012. PMID 16677733.
- ^ Heylbroeck C, Balachandran S, Servant MJ, DeLuca C, Barber GN, Lin R, et al. (April 2000). "The IRF-3 transcription factor mediates Sendai virus-induced apoptosis". Journal of Virology. 74 (8): 3781–92. doi:10.1128/jvi.74.8.3781-3792.2000. PMC 111887. PMID 10729153.
- ^ Cantell K, Hirvonen S, Kauppinen HL, Myllylä G (1981). "[4] Production of interferon in human leukocytes from normal donors with the use of Sendai virus". Production of interferon in human leukocytes from normal donors with the use of Sendai virus. Methods in Enzymology. Vol. 78. pp. 29–38. doi:10.1016/0076-6879(81)78094-7. ISBN 9780121819781. PMID 6173603.
- ^ Miettinen M, Sareneva T, Julkunen I, Matikainen S (October 2001). "IFNs activate toll-like receptor gene expression in viral infections". Genes and Immunity. 2 (6): 349–55. doi:10.1038/sj.gene.6363791. PMID 11607792. S2CID 5819381.
- ^ a b c d e Lappalainen J, Rintahaka J, Kovanen PT, Matikainen S, Eklund KK (April 2013). "Intracellular RNA recognition pathway activates strong anti-viral response in human mast cells". Clinical and Experimental Immunology. 172 (1): 121–8. doi:10.1111/cei.12042. PMC 3719938. PMID 23480192.
- ^ a b Neerincx A, Lautz K, Menning M, Kremmer E, Zigrino P, Hösel M, et al. (August 2010). "A role for the human nucleotide-binding domain, leucine-rich repeat-containing family member NLRC5 in antiviral responses". The Journal of Biological Chemistry. 285 (34): 26223–32. doi:10.1074/jbc.M110.109736. PMC 2924034. PMID 20538593.
- ^ a b Seth RB, Sun L, Ea CK, Chen ZJ (September 2005). "Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3". Cell. 122 (5): 669–682. doi:10.1016/j.cell.2005.08.012. PMID 16125763. S2CID 11104354.
- ^ Leib D (2010-03-08). "Evaluation for RIG-I detects viral genomic RNA during negative-strand RNA virus infection". Faculty of 1000. doi:10.3410/f.2412956.2047054. S2CID 90712939.
- ^ Mikkelsen SS, Jensen SB, Chiliveru S, Melchjorsen J, Julkunen I, Gaestel M, et al. (April 2009). "RIG-I-mediated activation of p38 MAPK is essential for viral induction of interferon and activation of dendritic cells: dependence on TRAF2 and TAK1". The Journal of Biological Chemistry. 284 (16): 10774–10782. doi:10.1074/jbc.M807272200. PMC 2667765. PMID 19224920.
- ^ Gitlin L, Benoit L, Song C, Cella M, Gilfillan S, Holtzman MJ, et al. (January 2010). "Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo". PLOS Pathogens. 6 (1): e1000734. doi:10.1371/journal.ppat.1000734. PMC 2809771. PMID 20107606.
- ^ Servant MJ, Grandvaux N, tenOever BR, Duguay D, Lin R, Hiscott J (March 2003). "Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA". The Journal of Biological Chemistry. 278 (11): 9441–7. doi:10.1074/jbc.M209851200. PMID 12524442. S2CID 19096582.
- ^ Barnes BJ, Moore PA, Pitha PM (June 2001). "Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon alpha genes". The Journal of Biological Chemistry. 276 (26): 23382–90. doi:10.1074/jbc.M101216200. PMID 11303025. S2CID 26896371.
- ^ a b c d Hua J, Liao MJ, Rashidbaigi A (July 1996). "Cytokines induced by Sendai virus in human peripheral blood leukocytes". Journal of Leukocyte Biology. 60 (1): 125–8. doi:10.1002/jlb.60.1.125. PMID 8699116. S2CID 28976518.
- ^ a b c d Costas MA, Mella D, Criscuolo M, Díaz A, Finkielman S, Nahmod VE, et al. (December 1993). "Superinduction of mitogen-stimulated interferon-gamma production and other lymphokines by Sendai virus". Journal of Interferon Research. 13 (6): 407–12. doi:10.1089/jir.1993.13.407. PMID 8151134.
- ^ a b c d Zidovec S, Mazuran R (February 1999). "Sendai virus induces various cytokines in human peripheral blood leukocytes: different susceptibility of cytokine molecules to low pH". Cytokine. 11 (2): 140–3. doi:10.1006/cyto.1998.0411. PMID 10089135.
- ^ Nyman TA, Tölö H, Parkkinen J, Kalkkinen N (January 1998). "Identification of nine interferon-alpha subtypes produced by Sendai virus-induced human peripheral blood leucocytes". The Biochemical Journal. 329 (Pt 2): 295–302. doi:10.1042/bj3290295. PMC 1219044. PMID 9425112.
- ^ Zeng J, Fournier P, Schirrmacher V (May 2002). "Induction of interferon-alpha and tumor necrosis factor-related apoptosis-inducing ligand in human blood mononuclear cells by hemagglutinin-neuraminidase but not F protein of Newcastle disease virus". Virology. 297 (1): 19–30. doi:10.1006/viro.2002.1413. PMID 12083832.
- ^ Génin P, Lin R, Hiscott J, Civas A (2012). "Recruitment of histone deacetylase 3 to the interferon-A gene promoters attenuates interferon expression". PLOS ONE. 7 (6): e38336. Bibcode:2012PLoSO...738336G. doi:10.1371/journal.pone.0038336. PMC 3369917. PMID 22685561.
- ^ a b c d Mandhana R, Horvath CM (November 2018). "Sendai Virus Infection Induces Expression of Novel RNAs in Human Cells". Scientific Reports. 8 (1): 16815. Bibcode:2018NatSR...816815M. doi:10.1038/s41598-018-35231-8. PMC 6235974. PMID 30429577.
- ^ Milone MC, Fitzgerald-Bocarsly P (September 1998). "The mannose receptor mediates induction of IFN-alpha in peripheral blood dendritic cells by enveloped RNA and DNA viruses". Journal of Immunology. 161 (5): 2391–9. doi:10.4049/jimmunol.161.5.2391. PMID 9725235. S2CID 30673547.
- ^ Eloranta ML, Sandberg K, Ricciardi-Castagnoli P, Lindahl M, Alm GV (September 1997). "Production of interferon-alpha/beta by murine dendritic cell lines stimulated by virus and bacteria". Scandinavian Journal of Immunology. 46 (3): 235–41. doi:10.1046/j.1365-3083.1997.d01-120.x. PMID 9315110. S2CID 40570647.
- ^ a b c d e Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A (March 2007). "Autophagy-dependent viral recognition by plasmacytoid dendritic cells". Science. 315 (5817): 1398–401. Bibcode:2007Sci...315.1398L. CiteSeerX 10.1.1.657.3208. doi:10.1126/science.1136880. PMID 17272685. S2CID 11549012.
- ^ Izaguirre A, Barnes BJ, Amrute S, Yeow WS, Megjugorac N, Dai J, et al. (December 2003). "Comparative analysis of IRF and IFN-alpha expression in human plasmacytoid and monocyte-derived dendritic cells". Journal of Leukocyte Biology. 74 (6): 1125–38. doi:10.1189/jlb.0603255. PMID 12960254. S2CID 12030752.
- ^ "Conventional dendritic cells - Latest research and news | Nature". www.nature.com. Retrieved 4 February 2020.
- ^ Vremec D, Pooley J, Hochrein H, Wu L, Shortman K (March 2000). "CD4 and CD8 expression by dendritic cell subtypes in mouse thymus and spleen". Journal of Immunology. 164 (6): 2978–86. doi:10.4049/jimmunol.164.6.2978. PMID 10706685. S2CID 20588521.
- ^ a b c Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J, et al. (February 2010). "Quantitative proteomics reveals subset-specific viral recognition in dendritic cells". Immunity. 32 (2): 279–89. doi:10.1016/j.immuni.2010.01.013. PMID 20171123.
- ^ a b c Kiener R, Fleischmann M, Wiegand MA, Lemmermann NA, Schwegler C, Kaufmann C, et al. (August 2018). "Efficient Delivery of Human Cytomegalovirus T Cell Antigens by Attenuated Sendai Virus Vectors". Journal of Virology. 92 (15). doi:10.1128/JVI.00569-18. PMC 6052310. PMID 29769344.
- ^ a b Donnelly RP, Kotenko SV (August 2010). "Interferon-lambda: a new addition to an old family". Journal of Interferon & Cytokine Research. 30 (8): 555–64. doi:10.1089/jir.2010.0078. PMC 2925029. PMID 20712453.
- ^ a b Yin Z, Dai J, Deng J, Sheikh F, Natalia M, Shih T, et al. (September 2012). "Type III IFNs are produced by and stimulate human plasmacytoid dendritic cells". Journal of Immunology. 189 (6): 2735–45. doi:10.4049/jimmunol.1102038. PMC 3579503. PMID 22891284.
- ^ a b c d e Suzuki H, Kurooka M, Hiroaki Y, Fujiyoshi Y, Kaneda Y (April 2008). "Sendai virus F glycoprotein induces IL-6 production in dendritic cells in a fusion-independent manner". FEBS Letters. 582 (9): 1325–9. Bibcode:2008FEBSL.582.1325S. doi:10.1016/j.febslet.2008.03.011. PMID 18358837. S2CID 207607018.
- ^ a b Kawaguchi Y, Miyamoto Y, Inoue T, Kaneda Y (May 2009). "Efficient eradication of hormone-resistant human prostate cancers by inactivated Sendai virus particle". International Journal of Cancer. 124 (10): 2478–87. doi:10.1002/ijc.24234. PMID 19173282. S2CID 33289879.
- ^ a b Lin HY, Davis PJ, Thacore HR (December 1991). "Production of human interferon-beta by Sendai virus and poly(rI).poly(rC): inhibition by neomycin". Journal of Interferon Research. 11 (6): 365–9. doi:10.1089/jir.1991.11.365. PMID 1666117.
- ^ a b c Reinert LS, Harder L, Holm CK, Iversen MB, Horan KA, Dagnæs-Hansen F, et al. (April 2012). "TLR3 deficiency renders astrocytes permissive to herpes simplex virus infection and facilitates establishment of CNS infection in mice". The Journal of Clinical Investigation. 122 (4): 1368–76. doi:10.1172/JCI60893. PMC 3314467. PMID 22426207.
- ^ Ito Y, Hosaka Y (March 1983). "Component(s) of Sendai virus that can induce interferon in mouse spleen cells". Infection and Immunity. 39 (3): 1019–23. doi:10.1128/IAI.39.3.1019-1023.1983. PMC 348058. PMID 6301988.
- ^ a b c d Subramanian G, Kuzmanovic T, Zhang Y, Peter CB, Veleeparambil M, Chakravarti R, et al. (January 2018). "A new mechanism of interferon's antiviral action: Induction of autophagy, essential for paramyxovirus replication, is inhibited by the interferon stimulated gene, TDRD7". PLOS Pathogens. 14 (1): e1006877. doi:10.1371/journal.ppat.1006877. PMC 5806901. PMID 29381763.
- ^ Wetzel JL, Fensterl V, Sen GC (December 2014). "Sendai virus pathogenesis in mice is prevented by Ifit2 and exacerbated by interferon". Journal of Virology. 88 (23): 13593–601. doi:10.1128/JVI.02201-14. PMC 4248979. PMID 25231314.
- ^ Haller O, Kochs G (January 2011). "Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity". Journal of Interferon & Cytokine Research. 31 (1): 79–87. doi:10.1089/jir.2010.0076. PMID 21166595.
- ^ a b Peters K, Chattopadhyay S, Sen GC (April 2008). "IRF-3 activation by Sendai virus infection is required for cellular apoptosis and avoidance of persistence". Journal of Virology. 82 (7): 3500–8. doi:10.1128/JVI.02536-07. PMC 2268502. PMID 18216110.
- ^ a b Pirhonen J, Matikainen S, Julkunen I (November 2002). "Regulation of virus-induced IL-12 and IL-23 expression in human macrophages". Journal of Immunology. 169 (10): 5673–5678. doi:10.4049/jimmunol.169.10.5673. PMID 12421946.
- ^ a b Ittah M, Miceli-Richard C, Lebon P, Pallier C, Lepajolec C, Mariette X (2011). "Induction of B cell-activating factor by viral infection is a general phenomenon, but the types of viruses and mechanisms depend on cell type". Journal of Innate Immunity. 3 (2): 200–7. doi:10.1159/000321194. PMID 21051868. S2CID 6699971.
- ^ a b Johansson E, Domeika K, Berg M, Alm GV, Fossum C (February 2003). "Characterisation of porcine monocyte-derived dendritic cells according to their cytokine profile". Veterinary Immunology and Immunopathology. 91 (3–4): 183–97. doi:10.1016/s0165-2427(02)00310-0. PMID 12586481.
- ^ a b Li S, Nishikawa T, Kaneda Y (December 2017). "Inactivated Sendai virus particle upregulates cancer cell expression of intercellular adhesion molecule-1 and enhances natural killer cell sensitivity on cancer cells". Cancer Science. 108 (12): 2333–2341. doi:10.1111/cas.13408. PMC 5715349. PMID 28945328.
- ^ Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, Franchi L, et al. (December 2006). "Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA". The Journal of Biological Chemistry. 281 (48): 36560–8. doi:10.1074/jbc.M607594200. PMID 17008311. S2CID 23488241.
- ^ Park S, Juliana C, Hong S, Datta P, Hwang I, Fernandes-Alnemri T, et al. (October 2013). "The mitochondrial antiviral protein MAVS associates with NLRP3 and regulates its inflammasome activity". Journal of Immunology. 191 (8): 4358–66. doi:10.4049/jimmunol.1301170. PMC 3848201. PMID 24048902.
- ^ Subramanian N, Natarajan K, Clatworthy MR, Wang Z, Germain RN (April 2013). "The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation". Cell. 153 (2): 348–61. doi:10.1016/j.cell.2013.02.054. PMC 3632354. PMID 23582325.
- ^ Ryan LK, Diamond G (June 2017). "Modulation of Human β-Defensin-1 Production by Viruses". Viruses. 9 (6): 153. doi:10.3390/v9060153. PMC 5490828. PMID 28635669.
- ^ Ryan LK, Dai J, Yin Z, Megjugorac N, Uhlhorn V, Yim S, et al. (August 2011). "Modulation of human beta-defensin-1 (hBD-1) in plasmacytoid dendritic cells (PDC), monocytes, and epithelial cells by influenza virus, Herpes simplex virus, and Sendai virus and its possible role in innate immunity". Journal of Leukocyte Biology. 90 (2): 343–56. doi:10.1189/jlb.0209079. PMC 3133436. PMID 21551252.
- ^ Öhman T, Söderholm S, Paidikondala M, Lietzén N, Matikainen S, Nyman TA (June 2015). "Phosphoproteome characterization reveals that Sendai virus infection activates mTOR signaling in human epithelial cells". Proteomics. 15 (12): 2087–2097. doi:10.1002/pmic.201400586. PMID 25764225. S2CID 32610482.
- ^ a b c d Samal SK (2021), Vanniasinkam T, Tikoo SK, Samal SK (eds.), "Paramyxoviruses as Vaccine Vectors", Viral Vectors in Veterinary Vaccine Development, Cham: Springer International Publishing, pp. 113–139, doi:10.1007/978-3-030-51927-8_8, ISBN 978-3-030-51926-1, retrieved 2024-05-26
- ^ a b Nakanishi M, Otsu M (October 2012). "Development of Sendai virus vectors and their potential applications in gene therapy and regenerative medicine". Current Gene Therapy. 12 (5): 410–6. doi:10.2174/156652312802762518. PMC 3504922. PMID 22920683.
- ^ Russell CJ, Hurwitz JL (May 2021). "Sendai Virus-Vectored Vaccines That Express Envelope Glycoproteins of Respiratory Viruses". Viruses. 13 (6): 1023. doi:10.3390/v13061023. PMC 8230104. PMID 34072332.
- ^ a b c d e Kinoh H, Inoue M, Washizawa K, Yamamoto T, Fujikawa S, Tokusumi Y, et al. (July 2004). "Generation of a recombinant Sendai virus that is selectively activated and lyses human tumor cells expressing matrix metalloproteinases". Gene Therapy. 11 (14): 1137–45. doi:10.1038/sj.gt.3302272. PMID 15085175. S2CID 10376042.
- ^ Kinoh H, Inoue M (January 2008). "New cancer therapy using genetically-engineered oncolytic Sendai virus vector". Frontiers in Bioscience. 13 (13): 2327–34. doi:10.2741/2847. PMID 17981715. S2CID 25851804.
- ^ Tatsuta K, Tanaka S, Tajiri T, Shibata S, Komaru A, Ueda Y, et al. (February 2009). "Complete elimination of established neuroblastoma by synergistic action of gamma-irradiation and DCs treated with rSeV expressing interferon-beta gene". Gene Therapy. 16 (2): 240–51. doi:10.1038/gt.2008.161. PMID 18987675. S2CID 27976395.
- ^ a b c d e f Zimmermann M, Armeanu-Ebinger S, Bossow S, Lampe J, Smirnow I, Schenk A, et al. (2014). "Attenuated and protease-profile modified sendai virus vectors as a new tool for virotherapy of solid tumors". PLOS ONE. 9 (3): e90508. Bibcode:2014PLoSO...990508Z. doi:10.1371/journal.pone.0090508. PMC 3944018. PMID 24598703.
- ^ Tanaka Y, Araki K, Tanaka S, Miyagawa Y, Suzuki H, Kamide D, et al. (June 2019). "Oncolytic Sendai virus-induced tumor-specific immunoresponses suppress "simulated metastasis" of squamous cell carcinoma in an immunocompetent mouse model". Head & Neck. 41 (6): 1676–1686. doi:10.1002/hed.25642. PMID 30620422. S2CID 58561289.
- ^ a b Yonemitsu Y, Ueda Y, Kinoh H, Hasegawa M (January 2008). "Immunostimulatory virotherapy using recombinant Sendai virus as a new cancer therapeutic regimen". Frontiers in Bioscience. 13 (13): 1892–8. doi:10.2741/2809. PMID 17981677.
- ^ Tanaka Y, Araki K, Tanaka S, Miyagawa Y, Suzuki H, Kamide D, et al. (August 2019). "Sentinel Lymph Node-Targeted Therapy by Oncolytic Sendai Virus Suppresses Micrometastasis of Head and Neck Squamous Cell Carcinoma in an Orthotopic Nude Mouse Model". Molecular Cancer Therapeutics. 18 (8): 1430–1438. doi:10.1158/1535-7163.MCT-18-1372. PMID 31171582. S2CID 174812921.
- ^ Iwadate Y, Inoue M, Saegusa T, Tokusumi Y, Kinoh H, Hasegawa M, et al. (May 2005). "Recombinant Sendai virus vector induces complete remission of established brain tumors through efficient interleukin-2 gene transfer in vaccinated rats". Clinical Cancer Research. 11 (10): 3821–7. doi:10.1158/1078-0432.CCR-04-1485. PMID 15897582. S2CID 37657020.
- ^ Tanaka M, Shimbo T, Kikuchi Y, Matsuda M, Kaneda Y (April 2010). "Sterile alpha motif containing domain 9 is involved in death signaling of malignant glioma treated with inactivated Sendai virus particle (HVJ-E) or type I interferon". International Journal of Cancer. 126 (8): 1982–1991. doi:10.1002/ijc.24965. PMID 19830690. S2CID 3414189.
- ^ Qian M, Tan HM, Yu N, Wang T, Zhang Q (April 2018). "Inactivated Sendai Virus Induces ROS-dependent Apoptosis and Autophagy in Human Prostate Cancer Cells". Biomedical and Environmental Sciences. 31 (4): 280–289. Bibcode:2018BioES..31..280Q. doi:10.3967/bes2018.036. PMID 29773091.
- ^ a b c d e Kurooka M, Kaneda Y (January 2007). "Inactivated Sendai virus particles eradicate tumors by inducing immune responses through blocking regulatory T cells". Cancer Research. 67 (1): 227–36. doi:10.1158/0008-5472.CAN-06-1615. PMID 17210703.
- ^ a b c Fujihara A, Kurooka M, Miki T, Kaneda Y (January 2008). "Intratumoral injection of inactivated Sendai virus particles elicits strong antitumor activity by enhancing local CXCL10 expression and systemic NK cell activation". Cancer Immunology, Immunotherapy. 57 (1): 73–84. doi:10.1007/s00262-007-0351-y. PMC 11030187. PMID 17602226. S2CID 8779015.
- ^ Nishikawa T, Tung LY, Kaneda Y (December 2014). "Systemic administration of platelets incorporating inactivated Sendai virus eradicates melanoma in mice". Molecular Therapy. 22 (12): 2046–55. doi:10.1038/mt.2014.128. PMC 4429689. PMID 25023327.
- ^ Zhang Q, Yuan WF, Zhai GQ, Zhu SY, Xue ZF, Zhu HF, et al. (October 2012). "Inactivated Sendai virus suppresses murine melanoma growth by inducing host immune responses and down-regulating β-catenin expression". Biomedical and Environmental Sciences. 25 (5): 509–16. Bibcode:2012BioES..25..509Z. doi:10.3967/0895-3988.2012.05.003. PMID 23122307.
- ^ Saga K, Tamai K, Yamazaki T, Kaneda Y (February 2013). "Systemic administration of a novel immune-stimulatory pseudovirion suppresses lung metastatic melanoma by regionally enhancing IFN-γ production". Clinical Cancer Research. 19 (3): 668–79. doi:10.1158/1078-0432.CCR-12-1947. PMID 23251005. S2CID 14105282.
- ^ Wheelock EF, Dingle JH (September 1964). "Observations on the Repeated Administration of Viruses to a Patient With Acute Leukemia. A Preliminary Report". The New England Journal of Medicine. 271 (13): 645–51. doi:10.1056/NEJM196409242711302. PMID 14170843.
- ^ Zainutdinov SS, Kochneva GV, Netesov SV, Chumakov PM, Matveeva OV (July 2019). "Directed evolution as a tool for the selection of oncolytic RNA viruses with desired phenotypes". Oncolytic Virotherapy. 8: 9–26. doi:10.2147/ov.s176523. PMC 6636189. PMID 31372363.
- ^ a b c d e Zainutdinov SS, Tikunov AY, Matveeva OV, Netesov SV, Kochneva GV (August 2016). "Complete Genome Sequence of the Oncolytic Sendai virus Strain Moscow". Genome Announcements. 4 (4). doi:10.1128/genomeA.00818-16. PMC 4982289. PMID 27516510.
- ^ Treatment of advanced metastatic cancers with oncolytic Sendai virus, 20 April 2014, archived from the original on 2021-12-19, retrieved 2019-08-21
- ^ a b US 9526779, Matveeva O, Senina A, Slav V, Senin M, "Method for cancer immunotherapy and pharmaceutical compositions based on oncolytic non-pathogenic Sendai virus", issued 27 December 2016, assigned to Sendai Viralytics LLC
- ^ Kiyohara E, Tanemura A, Nishioka M, Yamada M, Tanaka A, Yokomi A, et al. (June 2020). "Intratumoral injection of hemagglutinating virus of Japan-envelope vector yielded an antitumor effect for advanced melanoma: a phase I/IIa clinical study". Cancer Immunology, Immunotherapy. 69 (6): 1131–1140. doi:10.1007/s00262-020-02509-8. PMC 11027698. PMID 32047956. S2CID 211074332.
- ^ a b Matveeva OV, Chumakov PM (November 2018). "Defects in interferon pathways as potential biomarkers of sensitivity to oncolytic viruses". Reviews in Medical Virology. 28 (6): e2008. doi:10.1002/rmv.2008. PMC 6906582. PMID 30209859.
- ^ a b Liang JX, Liang Y, Gao W (May 2016). "Clinicopathological and prognostic significance of sialyl Lewis X overexpression in patients with cancer: a meta-analysis". OncoTargets and Therapy. 9: 3113–25. doi:10.2147/ott.s102389. PMC 4888715. PMID 27307752.
- ^ a b Blanas A, Sahasrabudhe NM, Rodríguez E, van Kooyk Y, van Vliet SJ (2018-05-11). "Corrigendum: Fucosylated Antigens in Cancer: An Alliance Toward Tumor Progression, Metastasis, and Resistance to Chemotherapy". Frontiers in Oncology. 8: 150. doi:10.3389/fonc.2018.00150. PMC 5958677. PMID 29795807.
- ^ a b Fuster MM, Esko JD (July 2005). "The sweet and sour of cancer: glycans as novel therapeutic targets". Nature Reviews. Cancer. 5 (7): 526–542. doi:10.1038/nrc1649. PMID 16069816. S2CID 10330140.
- ^ a b Noguchi M, Sato N, Sugimori H, Mori K, Oshimi K (October 2001). "A minor E-selectin ligand, CD65, is critical for extravascular infiltration of acute myeloid leukemia cells". Leukemia Research. 25 (10): 847–53. doi:10.1016/s0145-2126(01)00036-4. PMID 11532516.
- ^ a b c d e "KEGG GLYCAN: G00111". www.genome.jp. Retrieved 2019-08-13.
- ^ a b Liang YJ, Ding Y, Levery SB, Lobaton M, Handa K, Hakomori SI (March 2013). "Differential expression profiles of glycosphingolipids in human breast cancer stem cells vs. cancer non-stem cells". Proceedings of the National Academy of Sciences of the United States of America. 110 (13): 4968–73. Bibcode:2013PNAS..110.4968L. doi:10.1073/pnas.1302825110. PMC 3612608. PMID 23479608.
- ^ a b c Chambers M. "ChemIDplus - 71833-57-3 - OWMXULOUTAAVIX-HNZIOFRCSA-N - Sialosylparagloboside - Similar structures search, synonyms, formulas, resource links, and other chemical information". chem.nlm.nih.gov. Retrieved 2019-08-13.
- ^ a b Okegawa T (2018). "Detection of Circulating Tumor Cells in Castration-Resistant Prostate Cancer". Hormone Therapy and Castration Resistance of Prostate Cancer. Singapore: Springer. pp. 299–305. doi:10.1007/978-981-10-7013-6_30. ISBN 978-981-10-7012-9.
- ^ a b c d e Hatano K, Miyamoto Y, Nonomura N, Kaneda Y (October 2011). "Expression of gangliosides, GD1a, and sialyl paragloboside is regulated by NF-κB-dependent transcriptional control of α2,3-sialyltransferase I, II, and VI in human castration-resistant prostate cancer cells". International Journal of Cancer. 129 (8): 1838–47. doi:10.1002/ijc.25860. PMID 21165949. S2CID 7765966.
- ^ a b Hamasaki H, Aoyagi M, Kasama T, Handa S, Hirakawa K, Taki T (January 1999). "GT1b in human metastatic brain tumors: GT1b as a brain metastasis-associated ganglioside". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 1437 (1): 93–9. doi:10.1016/s1388-1981(98)00003-1. PMID 9931455.
- ^ a b c "KEGG GLYCAN: G00116". www.genome.jp. Retrieved 2019-08-13.
- ^ a b c "KEGG GLYCAN: G00117". www.genome.jp. Retrieved 2019-08-13.
- ^ a b Vukelić Z, Kalanj-Bognar S, Froesch M, Bîndila L, Radić B, Allen M, et al. (May 2007). "Human gliosarcoma-associated ganglioside composition is complex and distinctive as evidenced by high-performance mass spectrometric determination and structural characterization". Glycobiology. 17 (5): 504–15. doi:10.1093/glycob/cwm012. PMID 17293353.
- ^ a b c d e Markwell MA, Portner A, Schwartz AL (February 1985). "An alternative route of infection for viruses: entry by means of the asialoglycoprotein receptor of a Sendai virus mutant lacking its attachment protein". Proceedings of the National Academy of Sciences of the United States of America. 82 (4): 978–82. Bibcode:1985PNAS...82..978M. doi:10.1073/pnas.82.4.978. PMC 397176. PMID 2983337.
- ^ a b c d Bitzer M, Lauer U, Baumann C, Spiegel M, Gregor M, Neubert WJ (July 1997). "Sendai virus efficiently infects cells via the asialoglycoprotein receptor and requires the presence of cleaved F0 precursor proteins for this alternative route of cell entry". Journal of Virology. 71 (7): 5481–6. doi:10.1128/JVI.71.7.5481-5486.1997. PMC 191789. PMID 9188621.
- ^ a b "ASGR1 protein expression summary - The Human Protein Atlas". www.proteinatlas.org. Retrieved 2020-01-14.
- ^ "ASGR2 protein expression summary - The Human Protein Atlas". www.proteinatlas.org. Retrieved 2020-01-14.
- ^ a b "Asialoglycoprotein receptor 1 expression". The Human Protein Atlas. February 4, 2023. Retrieved February 4, 2023.
- ^ "Expression of Lewis-related antigen and prognosis in stage I non-small cell lung cancer". Lung Cancer. 13 (1): 93. August 1995. doi:10.1016/0169-5002(95)90215-5. ISSN 0169-5002.
- ^ Yu CJ, Shih JY, Lee YC, Shun CT, Yuan A, Yang PC (January 2005). "Sialyl Lewis antigens: association with MUC5AC protein and correlation with post-operative recurrence of non-small cell lung cancer". Lung Cancer. 47 (1): 59–67. doi:10.1016/j.lungcan.2004.05.018. PMID 15603855.
- ^ Sterner E, Flanagan N, Gildersleeve JC (July 2016). "Perspectives on Anti-Glycan Antibodies Gleaned from Development of a Community Resource Database". ACS Chemical Biology. 11 (7): 1773–1783. doi:10.1021/acschembio.6b00244. PMC 4949583. PMID 27220698. S2CID 17515010.
- ^ a b Ariga T, Bhat S, Kanda T, Yamawaki M, Tai T, Kushi Y, et al. (April 1996). "Expression and localization of Lewis(x) glycolipids and GD1a ganglioside in human glioma cells". Glycoconjugate Journal. 13 (2): 135–145. doi:10.1007/BF00731487. PMID 8737237. S2CID 11831860.
- ^ Cuello HA, Ferreira GM, Gulino CA, Toledo AG, Segatori VI, Gabri MR (December 2020). "Terminally sialylated and fucosylated complex N-glycans are involved in the malignant behavior of high-grade glioma". Oncotarget. 11 (52): 4822–4835. doi:10.18632/oncotarget.27850. PMC 7779250. PMID 33447350.
- ^ Son MJ, Woolard K, Nam DH, Lee J, Fine HA (May 2009). "SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma". Cell Stem Cell. 4 (5): 440–452. doi:10.1016/j.stem.2009.03.003. PMC 7227614. PMID 19427293.
- ^ Mao XG, Zhang X, Xue XY, Guo G, Wang P, Zhang W, et al. (December 2009). "Brain Tumor Stem-Like Cells Identified by Neural Stem Cell Marker CD15". Translational Oncology. 2 (4): 247–257. doi:10.1593/tlo.09136. PMC 2781066. PMID 19956386.
- ^ Patru C, Romao L, Varlet P, Coulombel L, Raponi E, Cadusseau J, et al. (February 2010). "CD133, CD15/SSEA-1, CD34 or side populations do not resume tumor-initiating properties of long-term cultured cancer stem cells from human malignant glio-neuronal tumors". BMC Cancer. 10 (1): 66. doi:10.1186/1471-2407-10-66. PMC 2841664. PMID 20181261.
- ^ Ward RJ, Lee L, Graham K, Satkunendran T, Yoshikawa K, Ling E, et al. (June 2009). "Multipotent CD15+ cancer stem cells in patched-1-deficient mouse medulloblastoma". Cancer Research. 69 (11): 4682–4690. doi:10.1158/0008-5472.CAN-09-0342. PMID 19487286.
- ^ Read TA, Fogarty MP, Markant SL, McLendon RE, Wei Z, Ellison DW, et al. (February 2009). "Identification of CD15 as a marker for tumor-propagating cells in a mouse model of medulloblastoma". Cancer Cell. 15 (2): 135–147. doi:10.1016/j.ccr.2008.12.016. PMC 2664097. PMID 19185848.
- ^ Singh AR, Joshi S, Zulcic M, Alcaraz M, Garlich JR, Morales GA, et al. (2016-03-03). Castresana JS (ed.). "PI-3K Inhibitors Preferentially Target CD15+ Cancer Stem Cell Population in SHH Driven Medulloblastoma". PLOS ONE. 11 (3): e0150836. Bibcode:2016PLoSO..1150836S. doi:10.1371/journal.pone.0150836. PMC 4777592. PMID 26938241.
- ^ Fukuoka K, Narita N, Saijo N (May 1998). "Increased expression of sialyl Lewis(x) antigen is associated with distant metastasis in lung cancer patients: immunohistochemical study on bronchofiberscopic biopsy specimens". Lung Cancer. 20 (2): 109–116. doi:10.1016/s0169-5002(98)00016-6. PMID 9711529.
- ^ Nakamori S, Kameyama M, Imaoka S, Furukawa H, Ishikawa O, Sasaki Y, et al. (April 1997). "Involvement of carbohydrate antigen sialyl Lewis(x) in colorectal cancer metastasis". Diseases of the Colon and Rectum. 40 (4): 420–431. doi:10.1007/bf02258386. PMID 9106690. S2CID 24770173.
- ^ Nakagoe T, Sawai T, Tsuji T, Jibiki M, Nanashima A, Yamaguchi H, et al. (March 2001). "Circulating sialyl Lewis(x), sialyl Lewis(a), and sialyl Tn antigens in colorectal cancer patients: multivariate analysis of predictive factors for serum antigen levels". Journal of Gastroenterology. 36 (3): 166–172. doi:10.1007/s005350170124. PMID 11291879. S2CID 25161348.
- ^ Yamadera M, Shinto E, Tsuda H, Kajiwara Y, Naito Y, Hase K, et al. (January 2018). "Sialyl Lewisx expression at the invasive front as a predictive marker of liver recurrence in stage II colorectal cancer". Oncology Letters. 15 (1): 221–228. doi:10.3892/ol.2017.7340. PMC 5769389. PMID 29391881.
- ^ Trinchera M, Aronica A, Dall'Olio F (February 2017). "Selectin Ligands Sialyl-Lewis a and Sialyl-Lewis x in Gastrointestinal Cancers". Biology. 6 (1): 16. doi:10.3390/biology6010016. PMC 5372009. PMID 28241499.
- ^ Nakagoe T, Fukushima K, Sawai T, Tsuji T, Jibiki M, Nanashima A, et al. (January 2002). "Increased expression of sialyl Lewis(x) antigen as a prognostic factor in patients with stage 0, I, and II gastric cancer". Cancer Letters. 175 (2): 213–221. doi:10.1016/S0304-3835(01)00705-4. PMID 11741750.
- ^ Nakagoe T, Fukushima K, Itoyanagi N, Ikuta Y, Oka T, Nagayasu T, et al. (May 2002). "Expression of ABH/Lewis-related antigens as prognostic factors in patients with breast cancer". Journal of Cancer Research and Clinical Oncology. 128 (5): 257–264. doi:10.1007/s00432-002-0334-5. PMID 12029441. S2CID 24553989.
- ^ Jeschke U, Mylonas I, Shabani N, Kunert-Keil C, Schindlbeck C, Gerber B, et al. (May 2005). "Expression of sialyl lewis X, sialyl Lewis A, E-cadherin and cathepsin-D in human breast cancer: immunohistochemical analysis in mammary carcinoma in situ, invasive carcinomas and their lymph node metastasis". Anticancer Research. 25 (3A): 1615–1622. PMID 16033070.
- ^ Carrascal MA, Silva M, Ferreira JA, Azevedo R, Ferreira D, Silva AM, et al. (September 2018). "A functional glycoproteomics approach identifies CD13 as a novel E-selectin ligand in breast cancer". Biochimica et Biophysica Acta (BBA) - General Subjects. 1862 (9): 2069–2080. doi:10.1016/j.bbagen.2018.05.013. PMID 29777742. S2CID 29167912.
- ^ Dimitroff CJ, Lechpammer M, Long-Woodward D, Kutok JL (August 2004). "Rolling of human bone-metastatic prostate tumor cells on human bone marrow endothelium under shear flow is mediated by E-selectin". Cancer Research. 64 (15): 5261–5269. doi:10.1158/0008-5472.CAN-04-0691. PMID 15289332. S2CID 11632075.
- ^ Munkley J (March 2017). "Glycosylation is a global target for androgen control in prostate cancer cells". Endocrine-Related Cancer. 24 (3): R49–R64. doi:10.1530/erc-16-0569. PMID 28159857.
- ^ Idikio HA (November 1997). "Sialyl-Lewis-X, Gleason grade and stage in non-metastatic human prostate cancer". Glycoconjugate Journal. 14 (7): 875–877. doi:10.1023/a:1018502424487. PMID 9511995. S2CID 28112794.
- ^ Fujii Y, Yoshida M, Chien LJ, Kihara K, Kageyama Y, Yasukochi Y, et al. (2000). "Significance of carbohydrate antigen sialyl-Lewis X, sialyl-Lewis A, and possible unknown ligands to adhesion of human urothelial cancer cells to activated endothelium". Urologia Internationalis. 64 (3): 129–133. doi:10.1159/000030512. PMID 10859542. S2CID 23332254.
- ^ Liang JX, Liang Y, Gao W (2016). "Clinicopathological and prognostic significance of sialyl Lewis X overexpression in patients with cancer: a meta-analysis". OncoTargets and Therapy. 9: 3113–3125. doi:10.2147/OTT.S102389. PMC 4888715. PMID 27307752.
- ^ Macher BA, Beckstead JH (1990-01-01). "Distribution of VIM-2 and SSEA-1 glycoconjugate epitopes among human leukocytes and leukemia cells". Leukemia Research. 14 (2): 119–130. doi:10.1016/0145-2126(90)90040-G. PMID 1690317.
- ^ Majdic O, Bettelheim P, Stockinger H, Aberer W, Liszka K, Lutz D, et al. (May 1984). "M2, a novel myelomonocytic cell surface antigen and its distribution on leukemic cells". International Journal of Cancer. 33 (5): 617–623. doi:10.1002/ijc.2910330511. PMID 6724736. S2CID 20483491.
- ^ Noguchi M, Sato N, Sugimori H, Mori K, Oshimi K (October 2001). "A minor E-selectin ligand, CD65, is critical for extravascular infiltration of acute myeloid leukemia cells". Leukemia Research. 25 (10): 847–853. doi:10.1016/s0145-2126(01)00036-4. PMID 11532516.
- ^ Liang YJ, Ding Y, Levery SB, Lobaton M, Handa K, Hakomori SI (March 2013). "Differential expression profiles of glycosphingolipids in human breast cancer stem cells vs. cancer non-stem cells". Proceedings of the National Academy of Sciences of the United States of America. 110 (13): 4968–4973. Bibcode:2013PNAS..110.4968L. doi:10.1073/pnas.1302825110. PMC 3612608. PMID 23479608.
- ^ Chahlavi A, Rayman P, Richmond AL, Biswas K, Zhang R, Vogelbaum M, et al. (June 2005). "Glioblastomas induce T-lymphocyte death by two distinct pathways involving gangliosides and CD70". Cancer Research. 65 (12): 5428–5438. doi:10.1158/0008-5472.CAN-04-4395. PMID 15958592.
- ^ a b Hatano K, Miyamoto Y, Nonomura N, Kaneda Y (October 2011). "Expression of gangliosides, GD1a, and sialyl paragloboside is regulated by NF-κB-dependent transcriptional control of α2,3-sialyltransferase I, II, and VI in human castration-resistant prostate cancer cells". International Journal of Cancer. 129 (8): 1838–1847. doi:10.1002/ijc.25860. PMID 21165949. S2CID 7765966.
- ^ Hamasaki H, Aoyagi M, Kasama T, Handa S, Hirakawa K, Taki T (January 1999). "GT1b in human metastatic brain tumors: GT1b as a brain metastasis-associated ganglioside". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 1437 (1): 93–99. doi:10.1016/S1388-1981(98)00003-1. PMID 9931455.
- ^ a b Merritt WD, Sztein MB, Taylor B, Reaman GH (December 1991). "Immunoreactivity of leukemic lymphoblasts of T-cell and B-cell precursor origin with monoclonal anti-GD3 and anti-GM3 antibodies". Leukemia. 5 (12): 1087–1091. PMID 1774957.
- ^ Westrick MA, Lee WM, Macher BA (December 1983). "Isolation and characterization of gangliosides from chronic myelogenous leukemia cells". Cancer Research. 43 (12 Pt 1): 5890–5894. PMID 6580065.
- ^ Sterner E, Flanagan N, Gildersleeve JC (July 2016). "Perspectives on Anti-Glycan Antibodies Gleaned from Development of a Community Resource Database". ACS Chemical Biology. 11 (7): 1773–83. doi:10.1021/acschembio.6b00244. PMC 4949583. PMID 27220698.
- ^ Britten CJ, Bird MI (February 1997). "Chemical modification of an alpha 3-fucosyltransferase; definition of amino acid residues essential for enzyme activity". Biochimica et Biophysica Acta (BBA) - General Subjects. 1334 (1): 57–64. doi:10.1016/s0304-4165(96)00076-1. PMID 9042366.
- ^ de Vries T, Knegtel RM, Holmes EH, Macher BA (October 2001). "Fucosyltransferases: structure/function studies". Glycobiology. 11 (10): 119R–128R. doi:10.1093/glycob/11.10.119r. PMID 11588153.
- ^ a b Shetterly S, Jost F, Watson SR, Knegtel R, Macher BA, Holmes EH (August 2007). "Site-specific fucosylation of sialylated polylactosamines by alpha1,3/4-fucosyltransferases-V and -VI Is defined by amino acids near the N terminus of the catalytic domain". The Journal of Biological Chemistry. 282 (34): 24882–92. doi:10.1074/jbc.m702395200. PMID 17604274. S2CID 27689343.
- ^ Trinchera M, Aronica A, Dall'Olio F (February 2017). "Selectin Ligands Sialyl-Lewis a and Sialyl-Lewis x in Gastrointestinal Cancers". Biology. 6 (1): 16. doi:10.3390/biology6010016. PMC 5372009. PMID 28241499.
- ^ a b Ngamukote S, Yanagisawa M, Ariga T, Ando S, Yu RK (December 2007). "Developmental changes of glycosphingolipids and expression of glycogenes in mouse brains". Journal of Neurochemistry. 103 (6): 2327–41. doi:10.1111/j.1471-4159.2007.04910.x. PMID 17883393. S2CID 21405747.
- ^ a b Xu W, Kozak CA, Desnick RJ (April 1995). "Uroporphyrinogen-III synthase: molecular cloning, nucleotide sequence, expression of a mouse full-length cDNA, and its localization on mouse chromosome 7". Genomics. 26 (3): 556–62. doi:10.1016/0888-7543(95)80175-l. PMID 7607680.
- ^ Vandermeersch S, Vanbeselaere J, Delannoy CP, Drolez A, Mysiorek C, Guérardel Y, et al. (April 2015). "Accumulation of GD1α Ganglioside in MDA-MB-231 Breast Cancer Cells Expressing ST6GalNAc V". Molecules. 20 (4): 6913–24. doi:10.3390/molecules20046913. PMC 6272744. PMID 25913930.
- ^ Chandrasekaran EV, Xue J, Xia J, Locke RD, Patil SA, Neelamegham S, et al. (November 2011). "Mammalian sialyltransferase ST3Gal-II: its exchange sialylation catalytic properties allow labeling of sialyl residues in mucin-type sialylated glycoproteins and specific gangliosides". Biochemistry. 50 (44): 9475–87. doi:10.1021/bi200301w. PMC 3206213. PMID 21913655.
- ^ "Expression of TMPRSS2 in cancer - Summary - the Human Protein Atlas".
- ^ "RT4". ATCC. Retrieved 20 August 2021.
- ^ Hidalgo IJ, Raub TJ, Borchardt RT (March 1989). "Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability". Gastroenterology. 96 (3): 736–49. doi:10.1016/0016-5085(89)90897-4. PMID 2914637.
- ^ "Cell atlas - TMPRSS2 - The Human Protein Atlas". www.proteinatlas.org. Retrieved 2019-08-20.
- ^ a b Katopodis P, Anikin V, Randeva HS, Spandidos DA, Chatha K, Kyrou I, et al. (August 2020). "Pan‑cancer analysis of transmembrane protease serine 2 and cathepsin L that mediate cellular SARS‑CoV‑2 infection leading to COVID-19". International Journal of Oncology. 57 (2): 533–539. doi:10.3892/ijo.2020.5071. PMC 7307597. PMID 32468052.
- ^ "Expression of TPSB2 in cancer - Summary - The Human Protein Atlas". www.proteinatlas.org. Retrieved 2019-08-20.
- ^ a b "Cell atlas - TPSB2 - The Human Protein Atlas". www.proteinatlas.org. Retrieved 2019-08-20.
- ^ Nilsson G, Blom T, Kusche-Gullberg M, Kjellén L, Butterfield JH, Sundström C, et al. (May 1994). "Phenotypic characterization of the human mast-cell line HMC-1". Scandinavian Journal of Immunology. 39 (5): 489–98. doi:10.1111/j.1365-3083.1994.tb03404.x. PMID 8191224. S2CID 28014083.
- ^ Martin P, Papayannopoulou T (June 1982). "HEL cells: a new human erythroleukemia cell line with spontaneous and induced globin expression". Science. 216 (4551): 1233–5. Bibcode:1982Sci...216.1233M. doi:10.1126/science.6177045. PMID 6177045.
- ^ Xiao H, He M, Xie G, Liu Y, Zhao Y, Ye X, et al. (October 2019). "The release of tryptase from mast cells promote tumor cell metastasis via exosomes". BMC Cancer. 19 (1): 1015. doi:10.1186/s12885-019-6203-2. PMC 6819443. PMID 31664930.
- ^ a b "Expression of PLG in cancer - Summary - The Human Protein Atlas". www.proteinatlas.org. Retrieved 2019-08-20.
- ^ "Expression of F10 in cancer - Summary - The Human Protein Atlas". www.proteinatlas.org. Retrieved 2019-08-30.
- ^ a b c d e f g Bedsaul JR, Zaritsky LA, Zoon KC (November 2016). "Type I Interferon-Mediated Induction of Antiviral Genes and Proteins Fails to Protect Cells from the Cytopathic Effects of Sendai Virus Infection". Journal of Interferon & Cytokine Research. 36 (11): 652–665. doi:10.1089/jir.2016.0051. PMC 5105340. PMID 27508859.
- ^ a b Schock SN, Chandra NV, Sun Y, Irie T, Kitagawa Y, Gotoh B, et al. (April 2017). "Induction of necroptotic cell death by viral activation of the RIG-I or STING pathway". Cell Death and Differentiation. 24 (4): 615–625. doi:10.1038/cdd.2016.153. PMC 5384020. PMID 28060376.
- ^ Goto H, Ohta K, Matsumoto Y, Yumine N, Nishio M (September 2016). López S (ed.). "Evidence that Receptor Destruction by the Sendai Virus Hemagglutinin-Neuraminidase Protein Is Responsible for Homologous Interference". Journal of Virology. 90 (17): 7640–7646. doi:10.1128/JVI.01087-16. ISSN 0022-538X. PMC 4988132. PMID 27279623.
- ^ a b Miyara M, Chader D, Sage E, Sugiyama D, Nishikawa H, Bouvry D, et al. (2015-06-09). "Sialyl Lewis x (CD15s) identifies highly differentiated and most suppressive FOXP3 high regulatory T cells in humans". Proceedings of the National Academy of Sciences. 112 (23): 7225–7230. doi:10.1073/pnas.1508224112. ISSN 0027-8424. PMC 4466753. PMID 26015572.
- ^ a b Ohishi K, Ishikura A, Nishida S, Abo H, Nakatsukasa H, Kawashima H (2024-06-01). "Sialyl Lewis X Defines an Activated and Functional Regulatory T Cell Subpopulation in Mice". The Journal of Immunology. 212 (11): 1627–1638. doi:10.4049/jimmunol.2300349. ISSN 0022-1767. PMID 38639586.
- ^ Ebert O, Shinozaki K, Kournioti C, Park MS, García-Sastre A, Woo SL (May 2004). "Syncytia induction enhances the oncolytic potential of vesicular stomatitis virus in virotherapy for cancer". Cancer Research. 64 (9): 3265–70. doi:10.1158/0008-5472.CAN-03-3753. PMID 15126368.
- ^ Nakamori M, Fu X, Meng F, Jin A, Tao L, Bast RC, et al. (July 2003). "Effective therapy of metastatic ovarian cancer with an oncolytic herpes simplex virus incorporating two membrane fusion mechanisms". Clinical Cancer Research. 9 (7): 2727–33. PMID 12855653.
- ^ Altomonte J, Marozin S, Schmid RM, Ebert O (February 2010). "Engineered newcastle disease virus as an improved oncolytic agent against hepatocellular carcinoma". Molecular Therapy. 18 (2): 275–84. doi:10.1038/mt.2009.231. PMC 2839313. PMID 19809404.
- ^ Tai JA, Chang CY, Nishikawa T, Kaneda Y (August 2019). "Cancer immunotherapy using the Fusion gene of Sendai virus" (PDF). Cancer Gene Therapy. 27 (6): 498–508. doi:10.1038/s41417-019-0126-6. PMID 31383952. S2CID 199450913.
- ^ Bateman A, Bullough F, Murphy S, Emiliusen L, Lavillette D, Cosset FL, et al. (March 2000). "Fusogenic membrane glycoproteins as a novel class of genes for the local and immune-mediated control of tumor growth". Cancer Research. 60 (6): 1492–7. PMID 10749110.
- ^ Galanis E, Bateman A, Johnson K, Diaz RM, James CD, Vile R, et al. (May 2001). "Use of viral fusogenic membrane glycoproteins as novel therapeutic transgenes in gliomas". Human Gene Therapy. 12 (7): 811–21. doi:10.1089/104303401750148766. PMID 11339897.
- ^ Lin EH, Salon C, Brambilla E, Lavillette D, Szecsi J, Cosset FL, et al. (April 2010). "Fusogenic membrane glycoproteins induce syncytia formation and death in vitro and in vivo: a potential therapy agent for lung cancer". Cancer Gene Therapy. 17 (4): 256–65. doi:10.1038/cgt.2009.74. PMID 19893593. S2CID 11203950.
- ^ Kursunel MA, Esendagli G (June 2017). "Corrigendum to "The untold story of IFN-γ in cancer biology" [Cytokine Growth Factor Rev. 31 (2016) 73-81]". Cytokine & Growth Factor Reviews. 35: 97. doi:10.1016/j.cytogfr.2017.02.002. PMID 28258821.
- ^ a b Ikeda H, Old LJ, Schreiber RD (April 2002). "The roles of IFN gamma in protection against tumor development and cancer immunoediting". Cytokine & Growth Factor Reviews. 13 (2): 95–109. doi:10.1016/s1359-6101(01)00038-7. PMID 11900986.
- ^ Dunn GP, Bruce AT, Sheehan KC, Shankaran V, Uppaluri R, Bui JD, et al. (July 2005). "A critical function for type I interferons in cancer immunoediting". Nature Immunology. 6 (7): 722–9. doi:10.1038/ni1213. PMID 15951814. S2CID 20374688.
- ^ Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, et al. (December 2007). "Interferons at age 50: past, current and future impact on biomedicine". Nature Reviews. Drug Discovery. 6 (12): 975–90. doi:10.1038/nrd2422. PMC 7097588. PMID 18049472. S2CID 583709.
- ^ Albini A, Marchisone C, Del Grosso F, Benelli R, Masiello L, Tacchetti C, et al. (April 2000). "Inhibition of angiogenesis and vascular tumor growth by interferon-producing cells: A gene therapy approach". The American Journal of Pathology. 156 (4): 1381–93. doi:10.1016/S0002-9440(10)65007-9. PMC 1876903. PMID 10751362.
- ^ Yogeeswaran G, Salk PL (June 1981). "Metastatic potential is positively correlated with cell surface sialylation of cultured murine tumor cell lines". Science. 212 (4502): 1514–6. Bibcode:1981Sci...212.1514Y. doi:10.1126/science.7233237. PMID 7233237.
- ^ Passaniti A, Hart GW (June 1988). "Cell surface sialylation and tumor metastasis. Metastatic potential of B16 melanoma variants correlates with their relative numbers of specific penultimate oligosaccharide structures". The Journal of Biological Chemistry. 263 (16): 7591–603. doi:10.1016/S0021-9258(18)68540-0. PMID 3372501.
- ^ Bresalier RS, Rockwell RW, Dahiya R, Duh QY, Kim YS (February 1990). "Cell surface sialoprotein alterations in metastatic murine colon cancer cell lines selected in an animal model for colon cancer metastasis". Cancer Research. 50 (4): 1299–307. PMID 2297775.
- ^ Pearlstein E, Salk PL, Yogeeswaran G, Karpatkin S (July 1980). "Correlation between spontaneous metastatic potential, platelet-aggregating activity of cell surface extracts, and cell surface sialylation in 10 metastatic-variant derivatives of a rat renal sarcoma cell line". Proceedings of the National Academy of Sciences of the United States of America. 77 (7): 4336–9. Bibcode:1980PNAS...77.4336P. doi:10.1073/pnas.77.7.4336. PMC 349829. PMID 6933486.
- ^ Benedetto A, Elia G, Sala A, Belardelli F (January 1989). "Hyposialylation of high-molecular-weight membrane glycoproteins parallels the loss of metastatic potential in wheat-germ agglutinin-resistant Friend leukemia cells". International Journal of Cancer. 43 (1): 126–33. doi:10.1002/ijc.2910430124. PMID 2910824. S2CID 24704777.
- ^ Hsu CC, Lin TW, Chang WW, Wu CY, Lo WH, Wang PH, et al. (February 2005). "Soyasaponin-I-modified invasive behavior of cancer by changing cell surface sialic acids". Gynecologic Oncology. 96 (2): 415–22. doi:10.1016/j.ygyno.2004.10.010. PMID 15661230.
- ^ Chang WW, Yu CY, Lin TW, Wang PH, Tsai YC (March 2006). "Soyasaponin I decreases the expression of alpha2,3-linked sialic acid on the cell surface and suppresses the metastatic potential of B16F10 melanoma cells". Biochemical and Biophysical Research Communications. 341 (2): 614–9. doi:10.1016/j.bbrc.2005.12.216. PMID 16427612.
- ^ Chiang CH, Wang CH, Chang HC, More SV, Li WS, Hung WC (May 2010). "A novel sialyltransferase inhibitor AL10 suppresses invasion and metastasis of lung cancer cells by inhibiting integrin-mediated signaling". Journal of Cellular Physiology. 223 (2): 492–9. doi:10.1002/jcp.22068. PMID 20112294. S2CID 24218458.
- ^ a b Cohen M, Elkabets M, Perlmutter M, Porgador A, Voronov E, Apte RN, et al. (November 2010). "Sialylation of 3-methylcholanthrene-induced fibrosarcoma determines antitumor immune responses during immunoediting". Journal of Immunology. 185 (10): 5869–78. doi:10.4049/jimmunol.1001635. PMID 20956342. S2CID 45698975.
- ^ Bossart KN, Fusco DL, Broder CC (2013). "Paramyxovirus Entry". Viral Entry into Host Cells. Advances in Experimental Medicine and Biology. Vol. 790. New York, NY: Springer. pp. 95–127. doi:10.1007/978-1-4614-7651-1_6. ISBN 978-1-4614-7650-4. PMC 8782154. PMID 23884588.
- ^ a b c d Powell LD, Whiteheart SW, Hart GW (July 1987). "Cell surface sialic acid influences tumor cell recognition in the mixed lymphocyte reaction". Journal of Immunology. 139 (1): 262–70. doi:10.4049/jimmunol.139.1.262. PMID 2953814. S2CID 25762755.
- ^ Tyagarajan K, Forte JG, Townsend RR (January 1996). "Exoglycosidase purity and linkage specificity: assessment using oligosaccharide substrates and high-pH anion-exchange chromatography with pulsed amperometric detection". Glycobiology. 6 (1): 83–93. doi:10.1093/glycob/6.1.83. PMID 8991514.
- ^ Drzeniek R, Gauhe A (February 1970). "Differences in substrate specificity of myxovirus neuraminidases". Biochemical and Biophysical Research Communications. 38 (4): 651–6. doi:10.1016/0006-291x(70)90630-3. PMID 5443707.
- ^ a b Brostrom MA, Bruening G, Bankowski RA (December 1971). "Comparison of neuraminidases of paramyxoviruses with immunologically dissimilar hemagglutinins". Virology. 46 (3): 856–65. doi:10.1016/0042-6822(71)90086-9. PMID 4332979.
- ^ Jarahian M, Watzl C, Fournier P, Arnold A, Djandji D, Zahedi S, et al. (August 2009). "Activation of natural killer cells by newcastle disease virus hemagglutinin-neuraminidase". Journal of Virology. 83 (16): 8108–21. doi:10.1128/JVI.00211-09. PMC 2715740. PMID 19515783.
- ^ Arnon TI, Lev M, Katz G, Chernobrov Y, Porgador A, Mandelboim O (September 2001). "Recognition of viral hemagglutinins by NKp44 but not by NKp30". European Journal of Immunology. 31 (9): 2680–9. doi:10.1002/1521-4141(200109)31:9<2680::AID-IMMU2680>3.0.CO;2-A. PMID 11536166. S2CID 22437769.
- ^ a b c Schirrmacher V, Haas C, Bonifer R, Ertel C (July 1997). "Virus potentiation of tumor vaccine T-cell stimulatory capacity requires cell surface binding but not infection". Clinical Cancer Research. 3 (7): 1135–48. PMID 9815793.
- ^ Ueda Y, Hasegawa M, Yonemitsu Y (2009). "Sendai Virus for Cancer Immunotherapy". Viral Applications of Green Fluorescent Protein. Methods in Molecular Biology. Vol. 515. Totowa, NJ: Humana Press. pp. 299–308. doi:10.1007/978-1-59745-559-6_21. ISBN 978-1-934115-87-9. PMID 19378123.
- ^ Harada Y, Yonemitsu Y (June 2011). "Dramatic improvement of DC-based immunotherapy against various malignancies". Frontiers in Bioscience. 16 (6): 2233–2242. doi:10.2741/3850. PMID 21622173. S2CID 30195748.
- ^ Shibata S, Okano S, Yonemitsu Y, Onimaru M, Sata S, Nagata-Takeshita H, et al. (September 2006). "Induction of efficient antitumor immunity using dendritic cells activated by recombinant Sendai virus and its modulation by exogenous IFN-beta gene". Journal of Immunology. 177 (6): 3564–3576. doi:10.4049/jimmunol.177.6.3564. PMID 16951315. S2CID 20134438.
- ^ Okano S, Yonemitsu Y, Shirabe K, Kakeji Y, Maehara Y, Harada M, et al. (February 2011). "Provision of continuous maturation signaling to dendritic cells by RIG-I-stimulating cytosolic RNA synthesis of Sendai virus". Journal of Immunology. 186 (3): 1828–1839. doi:10.4049/jimmunol.0901641. PMID 21187441. S2CID 36653582.
- ^ a b Sugiyama M, Kakeji Y, Tsujitani S, Harada Y, Onimaru M, Yoshida K, et al. (March 2011). "Antagonism of VEGF by genetically engineered dendritic cells is essential to induce antitumor immunity against malignant ascites". Molecular Cancer Therapeutics. 10 (3): 540–549. doi:10.1158/1535-7163.MCT-10-0479. hdl:2324/25486. PMID 21209070. S2CID 37616710.
- ^ Yoneyama Y, Ueda Y, Akutsu Y, Matsunaga A, Shimada H, Kato T, et al. (March 2007). "Development of immunostimulatory virotherapy using non-transmissible Sendai virus-activated dendritic cells". Biochemical and Biophysical Research Communications. 355 (1): 129–135. doi:10.1016/j.bbrc.2007.01.132. PMID 17292867.
- ^ Komaru A, Ueda Y, Furuya A, Tanaka S, Yoshida K, Kato T, et al. (October 2009). "Sustained and NK/CD4+ T cell-dependent efficient prevention of lung metastasis induced by dendritic cells harboring recombinant Sendai virus". Journal of Immunology. 183 (7): 4211–4219. doi:10.4049/jimmunol.0803845. PMID 19734206. S2CID 39194525.
- ^ Kato T, Ueda Y, Kinoh H, Yoneyama Y, Matsunaga A, Komaru A, et al. (November 2010). "RIG-I helicase-independent pathway in sendai virus-activated dendritic cells is critical for preventing lung metastasis of AT6.3 prostate cancer". Neoplasia. 12 (11): 906–914. doi:10.1593/neo.10732. PMC 2978913. PMID 21076616.
- ^ Edgar LJ, Thompson AJ, Vartabedian VF, Kikuchi C, Woehl JL, Teijaro JR, et al. (September 2021). "Sialic Acid Ligands of CD28 Suppress Costimulation of T Cells". ACS Central Science. 7 (9): 1508–1515. doi:10.1021/acscentsci.1c00525. PMC 8461770. PMID 34584952.
- ^ Silva M, Silva Z, Marques G, Ferro T, Gonçalves M, Monteiro M, et al. (July 2016). "Sialic acid removal from dendritic cells improves antigen cross-presentation and boosts anti-tumor immune responses". Oncotarget. 7 (27): 41053–41066. doi:10.18632/oncotarget.9419. ISSN 1949-2553. PMC 5173042. PMID 27203391.
- ^ Balneger N, Cornelissen LA, Wassink M, Moons SJ, Boltje TJ, Bar-Ephraim YE, et al. (February 2022). "Sialic acid blockade in dendritic cells enhances CD8+ T cell responses by facilitating high-avidity interactions". Cellular and Molecular Life Sciences. 79 (2): 98. doi:10.1007/s00018-021-04027-x. ISSN 1420-682X. PMC 8799591. PMID 35089436.
- ^ a b c Hosoya N, Miura T, Kawana-Tachikawa A, Koibuchi T, Shioda T, Odawara T, et al. (March 2008). "Comparison between Sendai virus and adenovirus vectors to transduce HIV-1 genes into human dendritic cells". Journal of Medical Virology. 80 (3): 373–82. doi:10.1002/jmv.21052. PMID 18205221. S2CID 31337462.
- ^ Nagai Y, ed. (2014-01-31). Sendai virus vector: advantages and applications. Tokyo: Springer. ISBN 978-4-431-54556-9. OCLC 870271420.
- ^ Parks CL (2017). "Replication-Competent Viral Vectors for Vaccine Delivery". Human Vaccines. Elsevier. pp. 25–63. doi:10.1016/b978-0-12-802302-0.00001-7. ISBN 978-0-12-802302-0.
- ^ a b c Sakai Y, Kiyotani K, Fukumura M, Asakawa M, Kato A, Shioda T, et al. (August 1999). "Accommodation of foreign genes into the Sendai virus genome: sizes of inserted genes and viral replication". FEBS Letters. 456 (2): 221–6. Bibcode:1999FEBSL.456..221S. doi:10.1016/s0014-5793(99)00960-6. PMID 10456313. S2CID 1285541.
- ^ a b Chare ER, Gould EA, Holmes EC (October 2003). "Phylogenetic analysis reveals a low rate of homologous recombination in negative-sense RNA viruses". The Journal of General Virology. 84 (Pt 10): 2691–2703. doi:10.1099/vir.0.19277-0. PMID 13679603.
- ^ a b Han GZ, Worobey M (August 2011). "Homologous recombination in negative sense RNA viruses". Viruses. 3 (8): 1358–1373. doi:10.3390/v3081358. PMC 3185808. PMID 21994784.
- ^ a b c Kolakofsky D, Roux L, Garcin D, Ruigrok RW (July 2005). "Paramyxovirus mRNA editing, the "rule of six" and error catastrophe: a hypothesis". The Journal of General Virology. 86 (Pt 7): 1869–1877. doi:10.1099/vir.0.80986-0. PMID 15958664.
- ^ a b Englund JA, Moscona A (2014), Kaslow RA, Stanberry LR, Le Duc JW (eds.), "Paramyxoviruses: Parainfluenza Viruses", Viral Infections of Humans, Boston, MA: Springer US, pp. 579–600, doi:10.1007/978-1-4899-7448-8_25, ISBN 978-1-4899-7447-1
- ^ a b Ikegame S, Beaty SM, Stevens C, Won T, Park A, Sachs D, et al. (October 2020). Lowen AC (ed.). "Genome-wide transposon mutagenesis of paramyxoviruses reveals constraints on genomic plasticity". PLOS Pathogens. 16 (10): e1008877. doi:10.1371/journal.ppat.1008877. PMC 7577504. PMID 33035269.
- ^ Garcin D, Pelet T, Calain P, Roux L, Curran J, Kolakofsky D (December 1995). "A highly recombinogenic system for the recovery of infectious Sendai paramyxovirus from cDNA: generation of a novel copy-back nondefective interfering virus". The EMBO Journal. 14 (24): 6087–94. doi:10.1002/j.1460-2075.1995.tb00299.x. PMC 394733. PMID 8557028.
- ^ Pfaller CK, Cattaneo R, Schnell MJ (May 2015). "Reverse genetics of Mononegavirales: How they work, new vaccines, and new cancer therapeutics". Virology. 60th Anniversary Issue. 479: 331–344. doi:10.1016/j.virol.2015.01.029. PMC 4557643. PMID 25702088.
- ^ Beaty SM, Park A, Won ST, Hong P, Lyons M, Vigant F, et al. (March 2017). "Paramyxoviridae Reverse Genetics Systems". mSphere. 2 (2). doi:10.1128/mSphere.00376-16. PMC 5371697. PMID 28405630.
- ^ Liu H, Albina E, Gil P, Minet C, de Almeida RS (September 2017). "Two-plasmid system to increase the rescue efficiency of paramyxoviruses by reverse genetics: The example of rescuing Newcastle Disease Virus". Virology. 509: 42–51. doi:10.1016/j.virol.2017.06.003. PMID 28595094.
- ^ Liu H, de Almeida RS, Gil P, Albina E (November 2017). "Comparison of the efficiency of different newcastle disease virus reverse genetics systems". Journal of Virological Methods. 249: 111–116. doi:10.1016/j.jviromet.2017.08.024. PMID 28867302.
- ^ Bajimaya S, Hayashi T, Takimoto T (2017). "Rescue of Sendai Virus from Cloned cDNA". Reverse Genetics of RNA Viruses. Methods in Molecular Biology. Vol. 1602. New York, NY: Springer. pp. 103–110. doi:10.1007/978-1-4939-6964-7_7. ISBN 978-1-4939-6962-3. PMID 28508216.
- ^ Yoshizaki M, Hironaka T, Iwasaki H, Ban H, Tokusumi Y, Iida A, et al. (September 2006). "Naked Sendai virus vector lacking all of the envelope-related genes: reduced cytopathogenicity and immunogenicity". The Journal of Gene Medicine. 8 (9): 1151–1159. doi:10.1002/jgm.938. PMID 16841365. S2CID 39942120.
- ^ Inoue M, Tokusumi Y, Ban H, Kanaya T, Shirakura M, Tokusumi T, et al. (June 2003). "A new Sendai virus vector deficient in the matrix gene does not form virus particles and shows extensive cell-to-cell spreading". Journal of Virology. 77 (11): 6419–6429. doi:10.1128/JVI.77.11.6419-6429.2003. PMC 155001. PMID 12743299.
- ^ a b c d e f Mostafa HH, Vogel P, Srinivasan A, Russell CJ (September 2016). "Non-invasive Imaging of Sendai Virus Infection in Pharmacologically Immunocompromised Mice: NK and T Cells, but not Neutrophils, Promote Viral Clearance after Therapy with Cyclophosphamide and Dexamethasone". PLOS Pathogens. 12 (9): e1005875. doi:10.1371/journal.ppat.1005875. PMC 5010285. PMID 27589232.
- ^ a b c d Hasan MK, Kato A, Shioda T, Sakai Y, Yu D, Nagai Y (November 1997). "Creation of an infectious recombinant Sendai virus expressing the firefly luciferase gene from the 3' proximal first locus". The Journal of General Virology. 78 (11): 2813–20. doi:10.1099/0022-1317-78-11-2813. PMID 9367367.
- ^ a b c d e f g Chambers R, Takimoto T (June 2010). "Trafficking of Sendai virus nucleocapsids is mediated by intracellular vesicles". PLOS ONE. 5 (6): e10994. Bibcode:2010PLoSO...510994C. doi:10.1371/journal.pone.0010994. PMC 2881874. PMID 20543880.
- ^ a b c Agungpriyono DR, Yamaguchi R, Uchida K, Tohya Y, Kato A, Nagai Y, et al. (February 2000). "Green fluorescent protein gene insertion of Sendai Virus infection in nude mice: possibility as an infection tracer". The Journal of Veterinary Medical Science. 62 (2): 223–8. doi:10.1292/jvms.62.223. PMID 10720198.
- ^ a b c Miyazaki M, Segawa H, Yamashita T, Zhu Y, Takizawa K, Hasegawa M, et al. (2010-11-23). "Construction and characterization of a fluorescent sendai virus carrying the gene for envelope fusion protein fused with enhanced green fluorescent protein". Bioscience, Biotechnology, and Biochemistry. 74 (11): 2293–8. doi:10.1271/bbb.100511. PMID 21071846. S2CID 43669142.
- ^ Touzelet O, Loukili N, Pelet T, Fairley D, Curran J, Power UF (March 2009). "De novo generation of a non-segmented negative strand RNA virus with a bicistronic gene". Virus Research. 140 (1–2): 40–48. doi:10.1016/j.virusres.2008.10.019. PMID 19084562.
- ^ a b Strähle L, Marq JB, Brini A, Hausmann S, Kolakofsky D, Garcin D (November 2007). "Activation of the beta interferon promoter by unnatural Sendai virus infection requires RIG-I and is inhibited by viral C proteins". Journal of Virology. 81 (22): 12227–37. doi:10.1128/JVI.01300-07. PMC 2169027. PMID 17804509.
- ^ a b Gosselin-Grenet AS, Mottet-Osman G, Roux L (September 2010). "Sendai virus particle production: basic requirements and role of the SYWST motif present in HN cytoplasmic tail". Virology. 405 (2): 439–447. doi:10.1016/j.virol.2010.06.030. PMID 20633915.
- ^ a b Rawling J, Cano O, Garcin D, Kolakofsky D, Melero JA (March 2011). "Recombinant Sendai viruses expressing fusion proteins with two furin cleavage sites mimic the syncytial and receptor-independent infection properties of respiratory syncytial virus". Journal of Virology. 85 (6): 2771–80. doi:10.1128/jvi.02065-10. PMC 3067931. PMID 21228237.
- ^ a b Rawling J, García-Barreno B, Melero JA (June 2008). "Insertion of the two cleavage sites of the respiratory syncytial virus fusion protein in Sendai virus fusion protein leads to enhanced cell-cell fusion and a decreased dependency on the HN attachment protein for activity". Journal of Virology. 82 (12): 5986–98. doi:10.1128/jvi.00078-08. PMC 2395136. PMID 18385247.
- ^ a b Portner A, Scroggs RA, Naeve CW (April 1987). "The fusion glycoprotein of Sendai virus: sequence analysis of an epitope involved in fusion and virus neutralization". Virology. 157 (2): 556–9. doi:10.1016/0042-6822(87)90301-1. PMID 2435061.
- ^ Park A, Hong P, Won ST, Thibault PA, Vigant F, Oguntuyo KY, et al. (2016-08-24). "Sendai virus, an RNA virus with no risk of genomic integration, delivers CRISPR/Cas9 for efficient gene editing". Molecular Therapy: Methods & Clinical Development. 3: 16057. doi:10.1038/mtm.2016.57. PMC 4996130. PMID 27606350.
- ^ a b c Corral T, Ver LS, Mottet G, Cano O, García-Barreno B, Calder LJ, et al. (April 2007). "High level expression of soluble glycoproteins in the allantoic fluid of embryonated chicken eggs using a Sendai virus minigenome system". BMC Biotechnology. 7 (1): 17. doi:10.1186/1472-6750-7-17. PMC 1852797. PMID 17411439.
- ^ Grygoryev D, Ekstrom T, Manalo E, Link JM, Alshaikh A, Keith D, et al. (March 2024). "Sendai virus is robust and consistent in delivering genes into human pancreatic cancer cells". Heliyon. 10 (5): e27221. Bibcode:2024Heliy..1027221G. doi:10.1016/j.heliyon.2024.e27221. PMC 10923719. PMID 38463758.
- ^ Fujie Y, Fusaki N, Katayama T, Hamasaki M, Soejima Y, Soga M, et al. (2014-12-05). "New type of Sendai virus vector provides transgene-free iPS cells derived from chimpanzee blood". PLOS ONE. 9 (12): e113052. Bibcode:2014PLoSO...9k3052F. doi:10.1371/journal.pone.0113052. PMC 4257541. PMID 25479600.
- ^ Jin CH, Kusuhara K, Yonemitsu Y, Nomura A, Okano S, Takeshita H, et al. (February 2003). "Recombinant Sendai virus provides a highly efficient gene transfer into human cord blood-derived hematopoietic stem cells". Gene Therapy. 10 (3): 272–7. doi:10.1038/sj.gt.3301877. PMID 12571635. S2CID 22415369.
- ^ Okumura T, Horie Y, Lai CY, Lin HT, Shoda H, Natsumoto B, et al. (June 2019). "Robust and highly efficient hiPSC generation from patient non-mobilized peripheral blood-derived CD34+ cells using the auto-erasable Sendai virus vector". Stem Cell Research & Therapy. 10 (1): 185. doi:10.1186/s13287-019-1273-2. PMC 6591940. PMID 31234949.
- ^ Seki T, Yuasa S, Fukuda K (March 2012). "Generation of induced pluripotent stem cells from a small amount of human peripheral blood using a combination of activated T cells and Sendai virus". Nature Protocols. 7 (4): 718–28. doi:10.1038/nprot.2012.015. PMID 22422317. S2CID 41397031.
- ^ "CTS CytoTune-iPS 2.1 Sendai Reprogramming Kit - Thermo Fisher Scientific". www.thermofisher.com. Retrieved 2019-08-13.
- ^ Charlesworth CT, Nakauchi H (November 2022). "An optimized Sendai viral vector platform for reprogramming to naive pluripotency". Cell Reports Methods. 2 (11): 100349. doi:10.1016/j.crmeth.2022.100349. PMC 9701616. PMID 36452874.
- ^ Sano M, Nakasu A, Ohtaka M, Nakanishi M (December 2019). "A Sendai Virus-Based Cytoplasmic RNA Vector as a Novel Platform for Long-Term Expression of MicroRNAs". Molecular Therapy. Methods & Clinical Development. 15: 371–382. doi:10.1016/j.omtm.2019.10.012. PMC 6889074. PMID 31828179.
- ^ Moriya C, Horiba S, Kurihara K, Kamada T, Takahara Y, Inoue M, et al. (November 2011). "Intranasal Sendai viral vector vaccination is more immunogenic than intramuscular under pre-existing anti-vector antibodies". Vaccine. 29 (47): 8557–8563. doi:10.1016/j.vaccine.2011.09.028. PMID 21939708.
- ^ a b Jones BG, Sealy RE, Rudraraju R, Traina-Dorge VL, Finneyfrock B, Cook A, et al. (January 2012). "Sendai virus-based RSV vaccine protects African green monkeys from RSV infection". Vaccine. 30 (5): 959–68. doi:10.1016/j.vaccine.2011.11.046. PMC 3256274. PMID 22119594.
- ^ Zhan X, Slobod KS, Jones BG, Sealy RE, Takimoto T, Boyd K, et al. (May 2015). "Sendai virus recombinant vaccine expressing a secreted, unconstrained respiratory syncytial virus fusion protein protects against RSV in cotton rats". International Immunology. 27 (5): 229–36. doi:10.1093/intimm/dxu107. PMC 4406265. PMID 25477211.
- ^ a b c d Hu Z, Gu L, Li CL, Shu T, Lowrie DB, Fan XY (2018). "The Profile of T Cell Responses in Bacille Calmette-Guérin-Primed Mice Boosted by a Novel Sendai Virus Vectored Anti-Tuberculosis Vaccine". Frontiers in Immunology. 9: 1796. doi:10.3389/fimmu.2018.01796. PMC 6085409. PMID 30123219.
- ^ Hu Z, Xia J, Wu J, Zhao H, Ji P, Gu L, et al. (December 2024). "A multistage Sendai virus vaccine incorporating latency-associated antigens induces protection against acute and latent tuberculosis". Emerging Microbes & Infections. 13 (1): 2300463. doi:10.1080/22221751.2023.2300463. ISSN 2222-1751. PMC 10769537. PMID 38164736.
- ^ Moreno-Fierros L, García-Silva I, Rosales-Mendoza S (August 2020). "Development of SARS-CoV-2 vaccines: should we focus on mucosal immunity?". Expert Opinion on Biological Therapy. 20 (8): 831–836. doi:10.1080/14712598.2020.1767062. PMID 32380868. S2CID 218556295.
- ^ Travis CR (2020-09-30). "As Plain as the Nose on Your Face: The Case for A Nasal (Mucosal) Route of Vaccine Administration for Covid-19 Disease Prevention". Frontiers in Immunology. 11: 591897. doi:10.3389/fimmu.2020.591897. PMC 7561361. PMID 33117404.
- ^ Sealy R, Jones BG, Surman SL, Hurwitz JL (September 2010). "Robust IgA and IgG-producing antibody forming cells in the diffuse-NALT and lungs of Sendai virus-vaccinated cotton rats associate with rapid protection against human parainfluenza virus-type 1". Vaccine. 28 (41): 6749–6756. doi:10.1016/j.vaccine.2010.07.068. PMC 2950074. PMID 20682364.
- ^ a b c Hu Z, Wong KW, Zhao HM, Wen HL, Ji P, Ma H, et al. (May 2017). "Sendai Virus Mucosal Vaccination Establishes Lung-Resident Memory CD8T Cell Immunity and Boosts BCG-Primed Protection against TB in Mice". Molecular Therapy. 25 (5): 1222–1233. doi:10.1016/j.ymthe.2017.02.018. PMC 5417795. PMID 28342639.
- ^ a b Yang Y, Ren L, Dong X, Wu X (2005-05-01). "108. The Complete Nucleotide Sequence of Sendai Virus Isolate BB1 and Comparison with Other Isolates". Molecular Therapy. 11: S44. doi:10.1016/j.ymthe.2005.06.469.
- ^ a b c d Shi LY, Li M, Yuan LJ, Wang Q, Li XM (2008). "A new paramyxovirus, Tianjin strain, isolated from common cotton-eared marmoset: genome characterization and structural protein sequence analysis". Archives of Virology. 153 (9): 1715–23. doi:10.1007/s00705-008-0184-9. PMID 18696006. S2CID 6151471.
- ^ Liang Y, Wu X, Zhang J, Xiao L, Yang Y, Bai X, et al. (December 2012). "Immunogenicity and therapeutic effects of Ag85A/B chimeric DNA vaccine in mice infected with Mycobacterium tuberculosis". FEMS Immunology and Medical Microbiology. 66 (3): 419–426. doi:10.1111/1574-695X.12008. PMID 23163873.
- ^ Ishii H, Nomura T, Yamamoto H, Nishizawa M, Thu Hau TT, Harada S, et al. (February 2022). "Neutralizing-antibody-independent SARS-CoV-2 control correlated with intranasal-vaccine-induced CD8+ T cell responses". Cell Reports. Medicine. 3 (2): 100520. doi:10.1016/j.xcrm.2022.100520. PMC 8768424. PMID 35233545.
- ^ "HN - Hemagglutinin-neuraminidase - Sendai virus (strain Z) (SeV) - HN gene & protein". www.uniprot.org. Retrieved 2019-08-09.
- ^ Scheid A, Choppin PW (February 1974). "Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis, and infectivity of proteolytic cleavage of an inactive precursor protein of Sendai virus". Virology. 57 (2): 475–90. doi:10.1016/0042-6822(74)90187-1. PMID 4361457.
- ^ "F - Fusion glycoprotein F0 precursor - Sendai virus (strain Z) (SeV) - F gene & protein". www.uniprot.org. Retrieved 2019-08-09.
- ^ a b c d Azarm KD, Lee B (January 2020). "Differential Features of Fusion Activation within the Paramyxoviridae". Viruses. 12 (2): 161. doi:10.3390/v12020161. PMC 7077268. PMID 32019182.
- ^ "M - Matrix protein - Sendai virus (strain Ohita) (SeV) - M gene & protein". www.uniprot.org. Retrieved 2019-08-09.
- ^ "N - Nucleoprotein - Sendai virus (strain Z) (SeV) - N gene & protein". www.uniprot.org. Retrieved 2019-08-09.
- ^ "P/V/C - Phosphoprotein - Sendai virus (strain Harris) (SeV) - P/V/C gene & protein". www.uniprot.org. Retrieved 2019-08-09.
- ^ "L - RNA-directed RNA polymerase L - Sendai virus (strain Enders) (SeV) - L gene & protein". www.uniprot.org. Retrieved 2019-08-09.
- ^ a b Yamada H, Hayata S, Omata-Yamada T, Taira H, Mizumoto K, Iwasaki K (1990). "Association of the Sendai virus C protein with nucleocapsids". Archives of Virology. 113 (3–4): 245–53. doi:10.1007/bf01316677. PMID 2171459. S2CID 24592567.
- ^ a b c d Garcin D, Curran J, Itoh M, Kolakofsky D (August 2001). "Longer and shorter forms of Sendai virus C proteins play different roles in modulating the cellular antiviral response". Journal of Virology. 75 (15): 6800–7. doi:10.1128/JVI.75.15.6800-6807.2001. PMC 114406. PMID 11435558.
- ^ a b c Curran J, Kolakofsky D (January 1988). "Ribosomal initiation from an ACG codon in the Sendai virus P/C mRNA". The EMBO Journal. 7 (1): 245–51. doi:10.1002/j.1460-2075.1988.tb02806.x. PMC 454264. PMID 2834203.
- ^ a b Dillon PJ, Gupta KC (February 1989). "Expression of five proteins from the Sendai virus P/C mRNA in infected cells". Journal of Virology. 63 (2): 974–7. doi:10.1128/JVI.63.2.974-977.1989. PMC 247778. PMID 2536120.
- ^ a b c de Breyne S, Simonet V, Pelet T, Curran J (January 2003). "Identification of a cis-acting element required for shunt-mediated translational initiation of the Sendai virus Y proteins". Nucleic Acids Research. 31 (2): 608–18. doi:10.1093/nar/gkg143. PMC 140508. PMID 12527769.
- ^ a b Curran J, Kolakofsky D (September 1988). "Scanning independent ribosomal initiation of the Sendai virus X protein". The EMBO Journal. 7 (9): 2869–74. doi:10.1002/j.1460-2075.1988.tb03143.x. PMC 457080. PMID 2846286.
- ^ Irie T, Nagata N, Yoshida T, Sakaguchi T (February 2008). "Recruitment of Alix/AIP1 to the plasma membrane by Sendai virus C protein facilitates budding of virus-like particles". Virology. 371 (1): 108–20. doi:10.1016/j.virol.2007.09.020. PMID 18028977.
- ^ a b Sakaguchi T, Kato A, Sugahara F, Shimazu Y, Inoue M, Kiyotani K, et al. (July 2005). "AIP1/Alix is a binding partner of Sendai virus C protein and facilitates virus budding". Journal of Virology. 79 (14): 8933–41. doi:10.1128/JVI.79.14.8933-8941.2005. PMC 1168738. PMID 15994787.
- ^ Payne V, Kam PC (July 2004). "Mast cell tryptase: a review of its physiology and clinical significance". Anaesthesia. 59 (7): 695–703. doi:10.1111/j.1365-2044.2004.03757.x. PMID 15200544. S2CID 7611291.
- ^ Chen Y, Shiota M, Ohuchi M, Towatari T, Tashiro J, Murakami M, et al. (June 2000). "Mast cell tryptase from pig lungs triggers infection by pneumotropic Sendai and influenza A viruses. Purification and characterization". European Journal of Biochemistry. 267 (11): 3189–97. doi:10.1046/j.1432-1327.2000.01346.x. PMID 10824103.
- ^ Tashiro M, Yokogoshi Y, Tobita K, Seto JT, Rott R, Kido H (December 1992). "Tryptase Clara, an activating protease for Sendai virus in rat lungs, is involved in pneumopathogenicity". Journal of Virology. 66 (12): 7211–6. doi:10.1128/JVI.66.12.7211-7216.1992. PMC 240423. PMID 1331518.
- ^ Kido H, Niwa Y, Beppu Y, Towatari T (1996). "Cellular proteases involved in the pathogenicity of enveloped animal viruses, human immunodeficiency virus, influenza virus A and Sendai virus". Advances in Enzyme Regulation. 36: 325–47. doi:10.1016/0065-2571(95)00016-X. PMID 8869754.
- ^ Le TQ, Kawachi M, Yamada H, Shiota M, Okumura Y, Kido H (April 2006). "Identification of trypsin I as a candidate for influenza A virus and Sendai virus envelope glycoprotein processing protease in rat brain". Biological Chemistry. 387 (4): 467–75. doi:10.1515/BC.2006.062. PMID 16606346. S2CID 11969821.
- ^ Murakami M, Towatari T, Ohuchi M, Shiota M, Akao M, Okumura Y, et al. (May 2001). "Mini-plasmin found in the epithelial cells of bronchioles triggers infection by broad-spectrum influenza A viruses and Sendai virus". European Journal of Biochemistry. 268 (10): 2847–55. doi:10.1046/j.1432-1327.2001.02166.x. PMID 11358500.
- ^ a b c Abe M, Tahara M, Sakai K, Yamaguchi H, Kanou K, Shirato K, et al. (November 2013). "TMPRSS2 is an activating protease for respiratory parainfluenza viruses". Journal of Virology. 87 (21): 11930–5. doi:10.1128/JVI.01490-13. PMC 3807344. PMID 23966399.
- ^ Gotoh B, Ogasawara T, Toyoda T, Inocencio NM, Hamaguchi M, Nagai Y (December 1990). "An endoprotease homologous to the blood clotting factor X as a determinant of viral tropism in chick embryo". The EMBO Journal. 9 (12): 4189–95. doi:10.1002/j.1460-2075.1990.tb07643.x. PMC 552195. PMID 2174359.
- ^ Ogasawara T, Gotoh B, Suzuki H, Asaka J, Shimokata K, Rott R, et al. (February 1992). "Expression of factor X and its significance for the determination of paramyxovirus tropism in the chick embryo". The EMBO Journal. 11 (2): 467–72. doi:10.1002/j.1460-2075.1992.tb05076.x. PMC 556476. PMID 1371460.
- ^ Gotoh B, Yamauchi F, Ogasawara T, Nagai Y (January 1992). "Isolation of factor Xa from chick embryo as the amniotic endoprotease responsible for paramyxovirus activation". FEBS Letters. 296 (3): 274–8. Bibcode:1992FEBSL.296..274G. doi:10.1016/0014-5793(92)80303-x. PMID 1537403. S2CID 33852517.
- ^ a b c d e f Lam A, Kirkland OO, Anderson PF, Seetharaman N, Vujovic D, Thibault PA, et al. (2021-11-23). "Single virus assay reveals membrane determinants and mechanistic features of Sendai virus binding". Biophysical Journal. 121 (6): 956–965. Bibcode:2022BpJ...121..956L. doi:10.1016/j.bpj.2022.02.011. PMC 8943810. PMID 35150620. S2CID 246715171.
- ^ a b c Matveeva OV, Shabalina SA (December 2020). "Prospects for Using Expression Patterns of Paramyxovirus Receptors as Biomarkers for Oncolytic Virotherapy". Cancers. 12 (12): 3659. doi:10.3390/cancers12123659. PMC 7762160. PMID 33291506.
- ^ a b c Villar E, Barroso IM (February 2006). "Role of sialic acid-containing molecules in paramyxovirus entry into the host cell: a minireview". Glycoconjugate Journal. 23 (1–2): 5–17. doi:10.1007/s10719-006-5433-0. PMID 16575518. S2CID 21083897.
- ^ a b Stockert RJ (July 1995). "The asialoglycoprotein receptor: relationships between structure, function, and expression". Physiological Reviews. 75 (3): 591–609. doi:10.1152/physrev.1995.75.3.591. PMID 7624395.
- ^ Suzuki Y, Suzuki T, Matsumoto M (June 1983). "Isolation and characterization of receptor sialoglycoprotein for hemagglutinating virus of Japan (Sendai virus) from bovine erythrocyte membrane". Journal of Biochemistry. 93 (6): 1621–33. doi:10.1093/oxfordjournals.jbchem.a134301. PMID 6309760.
- ^ a b Oku N, Nojima S, Inoue K (August 1981). "Studies on the interaction of Sendai virus with liposomal membranes. Sendai virus-induced agglutination of liposomes containing glycophorin". Biochimica et Biophysica Acta (BBA) - Biomembranes. 646 (1): 36–42. doi:10.1016/0005-2736(81)90269-8. PMID 6168285.
- ^ a b c d Müthing J (September 1996). "Influenza A and Sendai viruses preferentially bind to fucosylated gangliosides with linear poly-N-acetyllactosaminyl chains from human granulocytes". Carbohydrate Research. 290 (2): 217–24. doi:10.1016/0008-6215(96)00149-8. PMID 8823909.
- ^ "KEGG GLYCAN: G00197". www.genome.jp. Retrieved 2019-08-13.
- ^ Holmgren J, Svennerholm L, Elwing H, Fredman P, Strannegård O (April 1980). "Sendai virus receptor: proposed recognition structure based on binding to plastic-adsorbed gangliosides". Proceedings of the National Academy of Sciences of the United States of America. 77 (4): 1947–50. Bibcode:1980PNAS...77.1947H. doi:10.1073/pnas.77.4.1947. PMC 348626. PMID 6246515.
- ^ "Correction: Specific Gangliosides Function as Host Cell Receptors for Sendai Virus". Proceedings of the National Academy of Sciences. 79 (3): 951. 1982-02-01. Bibcode:1982PNAS...79Q.951.. doi:10.1073/pnas.79.3.951.
- ^ a b c Suzuki Y, Suzuki T, Matsunaga M, Matsumoto M (April 1985). "Gangliosides as paramyxovirus receptor. Structural requirement of sialo-oligosaccharides in receptors for hemagglutinating virus of Japan (Sendai virus) and Newcastle disease virus". Journal of Biochemistry. 97 (4): 1189–99. doi:10.1093/oxfordjournals.jbchem.a135164. PMID 2993261.
- ^ a b c d e f g h Suzuki T, Portner A, Scroggs RA, Uchikawa M, Koyama N, Matsuo K, et al. (May 2001). "Receptor specificities of human respiroviruses". Journal of Virology. 75 (10): 4604–13. doi:10.1128/JVI.75.10.4604-4613.2001. PMC 114213. PMID 11312330.
- ^ "KEGG GLYCAN: G00112". www.genome.jp. Retrieved 2019-08-13.
- ^ "KEGG GLYCAN: G00122". www.genome.jp. Retrieved 2019-08-13.
- ^ Umeda M, Nojima S, Inoue K (February 1984). "Activity of human erythrocyte gangliosides as a receptor to HVJ". Virology. 133 (1): 172–82. doi:10.1016/0042-6822(84)90436-7. PMID 6322427.
- ^ Suzuki T, Portner A, Scroggs RA, Uchikawa M, Koyama N, Matsuo K, et al. (May 2001). "Receptor specificities of human respiroviruses". Journal of Virology. 75 (10): 4604–13. doi:10.1128/jvi.75.10.4604-4613.2001. PMC 114213. PMID 11312330.
- ^ Mariethoz J, Khatib K, Campbell MP, Packer NH, Mullen E, Lisacek F (2014-10-20). "SugarBindDB: Resource of Pathogen Lectin-Glycan Interactions". Glycoscience: Biology and Medicine. Springer Japan. pp. 275–282. doi:10.1007/978-4-431-54841-6_28. ISBN 9784431548409.
- ^ "DBGET search - GLYCAN". www.genome.jp. Retrieved 2019-08-15.
- ^ "About PubChem". pubchemdocs.ncbi.nlm.nih.gov. Retrieved 2019-08-15.
- ^ "TOXNET". toxnet.nlm.nih.gov. Retrieved 2019-08-15.
- ^ a b Chang A, Dutch RE (April 2012). "Paramyxovirus fusion and entry: multiple paths to a common end". Viruses. 4 (4): 613–36. doi:10.3390/v4040613. PMC 3347325. PMID 22590688.
- ^ Lamb RA, Jardetzky TS (August 2007). "Structural basis of viral invasion: lessons from paramyxovirus F". Current Opinion in Structural Biology. 17 (4): 427–36. doi:10.1016/j.sbi.2007.08.016. PMC 2086805. PMID 17870467.
- ^ a b Haywood AM (November 2010). "Membrane uncoating of intact enveloped viruses". Journal of Virology. 84 (21): 10946–55. doi:10.1128/JVI.00229-10. PMC 2953184. PMID 20668081.
- ^ Jardetzky TS, Lamb RA (April 2014). "Activation of paramyxovirus membrane fusion and virus entry". Current Opinion in Virology. 5: 24–33. doi:10.1016/j.coviro.2014.01.005. PMC 4028362. PMID 24530984.
- ^ Whelan SP, Barr JN, Wertz GW (2004). "Transcription and Replication of Nonsegmented Negative-Strand RNA Viruses". Biology of Negative Strand RNA Viruses: The Power of Reverse Genetics. Current Topics in Microbiology and Immunology. Vol. 283. Berlin, Heidelberg: Springer. pp. 61–119. doi:10.1007/978-3-662-06099-5_3. ISBN 978-3-642-07375-5. PMID 15298168.
- ^ Noton SL, Fearns R (May 2015). "Initiation and regulation of paramyxovirus transcription and replication". Virology. 479: 545–354. doi:10.1016/j.virol.2015.01.014. PMC 4424093. PMID 25683441.
- ^ Ogino T, Kobayashi M, Iwama M, Mizumoto K (February 2005). "Sendai virus RNA-dependent RNA polymerase L protein catalyzes cap methylation of virus-specific mRNA". The Journal of Biological Chemistry. 280 (6): 4429–35. doi:10.1074/jbc.M411167200. PMID 15574411. S2CID 27655763.
- ^ a b Myers TM, Moyer SA (February 1997). "An amino-terminal domain of the Sendai virus nucleocapsid protein is required for template function in viral RNA synthesis". Journal of Virology. 71 (2): 918–24. doi:10.1128/JVI.71.2.918-924.1997. PMC 191139. PMID 8995608.
- ^ Ryu SW (2017). "Other Negative-Strand RNA Viruses". Molecular Virology of Human Pathogenic Viruses. Elsevier. pp. 213–224. doi:10.1016/b978-0-12-800838-6.00016-3. ISBN 978-0-12-800838-6. S2CID 88812845.
- ^ Wiegand MA, Bossow S, Schlecht S, Neubert WJ (2007-12-15). "De Novo Synthesis of N and P Proteins as a Key Step in Sendai Virus Gene Expression". Journal of Virology. 81 (24): 13835–13844. doi:10.1128/jvi.00914-07. ISSN 0022-538X. PMC 2168830.
- ^ Latorre P, Kolakofsky D, Curran J (September 1998). "Sendai virus Y proteins are initiated by a ribosomal shunt". Molecular and Cellular Biology. 18 (9): 5021–31. doi:10.1128/mcb.18.9.5021. PMC 109087. PMID 9710586.
- ^ a b c Harrison MS, Sakaguchi T, Schmitt AP (September 2010). "Paramyxovirus assembly and budding: building particles that transmit infections". The International Journal of Biochemistry & Cell Biology. 42 (9): 1416–1429. doi:10.1016/j.biocel.2010.04.005. PMC 2910131. PMID 20398786.
- ^ Miazza V, Mottet-Osman G, Startchick S, Chaponnier C, Roux L (February 2011). "Sendai virus induced cytoplasmic actin remodeling correlates with efficient virus particle production". Virology. 410 (1): 7–16. doi:10.1016/j.virol.2010.10.003. PMID 21075412.
- ^ a b c Ito M, Takeuchi T, Nishio M, Kawano M, Komada H, Tsurudome M, et al. (November 2004). "Early stage of establishment of persistent Sendai virus infection: unstable dynamic phase and then selection of viruses which are tightly cell associated, temperature sensitive, and capable of establishing persistent infection". Journal of Virology. 78 (21): 11939–11951. doi:10.1128/JVI.78.21.11939-11951.2004. PMC 523293. PMID 15479834.
- ^ Iwata M, Kawabata R, Morimoto N, Takeuchi RF, Sakaguchi T, Irie T, et al. (2024-04-02). "Evolutionary engineering and characterization of Sendai virus mutants capable of persistent infection and autonomous production". Frontiers in Virology. 4. doi:10.3389/fviro.2024.1363092. ISSN 2673-818X.
- ^ Coakley C, Peter C, Fabry S, Chattopadhyay S (August 2017). "Establishment of a Human Cell Line Persistently Infected with Sendai Virus". Bio-Protocol. 7 (16): e2512. doi:10.21769/BioProtoc.2512. PMC 8413619. PMID 34541174.
- ^ Chattopadhyay S, Fensterl V, Zhang Y, Veleeparambil M, Yamashita M, Sen GC (January 2013). "Role of interferon regulatory factor 3-mediated apoptosis in the establishment and maintenance of persistent infection by Sendai virus". Journal of Virology. 87 (1): 16–24. doi:10.1128/JVI.01853-12. PMC 3536409. PMID 23077293.
- ^ a b Mercado-López X, Cotter CR, Kim WK, Sun Y, Muñoz L, Tapia K, et al. (November 2013). "Highly immunostimulatory RNA derived from a Sendai virus defective viral genome". Vaccine. 31 (48): 5713–5721. doi:10.1016/j.vaccine.2013.09.040. PMC 4406099. PMID 24099876.
- ^ Xu J, Sun Y, Li Y, Ruthel G, Weiss SR, Raj A, et al. (October 2017). "Replication defective viral genomes exploit a cellular pro-survival mechanism to establish paramyxovirus persistence". Nature Communications. 8 (1): 799. Bibcode:2017NatCo...8..799X. doi:10.1038/s41467-017-00909-6. PMC 5630589. PMID 28986577.
- ^ a b c Genoyer E, López CB (February 2019). "Defective Viral Genomes Alter How Sendai Virus Interacts with Cellular Trafficking Machinery, Leading to Heterogeneity in the Production of Viral Particles among Infected Cells". Journal of Virology. 93 (4). doi:10.1128/JVI.01579-18. PMC 6364009. PMID 30463965.
- ^ a b c Pablo-Maiso L, Echeverría I, Rius-Rocabert S, Luján L, Garcin D, Andrés D, et al. (April 2020). "Sendai Virus, a Strong Inducer of Anti-Lentiviral State in Ovine Cells". Vaccines. 8 (2): 206. doi:10.3390/vaccines8020206. PMC 7349755. PMID 32365702.
- ^ Faísca P, Desmecht D (February 2007). "Sendai virus, the mouse parainfluenza type 1: a longstanding pathogen that remains up-to-date". Research in Veterinary Science. 82 (1): 115–125. doi:10.1016/j.rvsc.2006.03.009. PMID 16759680.
- ^ Rawling J, Cano O, Garcin D, Kolakofsky D, Melero JA (March 2011). "Recombinant Sendai viruses expressing fusion proteins with two furin cleavage sites mimic the syncytial and receptor-independent infection properties of respiratory syncytial virus". Journal of Virology. 85 (6): 2771–80. doi:10.1128/JVI.02065-10. PMC 3067931. PMID 21228237.
- ^ Hoekstra D, Klappe K, Hoff H, Nir S (April 1989). "Mechanism of fusion of Sendai virus: role of hydrophobic interactions and mobility constraints of viral membrane proteins. Effects of polyethylene glycol". The Journal of Biological Chemistry. 264 (12): 6786–92. doi:10.1016/S0021-9258(18)83498-6. PMID 2540161.
- ^ Takimoto T, Taylor GL, Connaris HC, Crennell SJ, Portner A (December 2002). "Role of the hemagglutinin-neuraminidase protein in the mechanism of paramyxovirus-cell membrane fusion". Journal of Virology. 76 (24): 13028–33. doi:10.1128/JVI.76.24.13028-13033.2002. PMC 136693. PMID 12438628.
- ^ Novick SL, Hoekstra D (October 1988). "Membrane penetration of Sendai virus glycoproteins during the early stages of fusion with liposomes as determined by hydrophobic photoaffinity labeling". Proceedings of the National Academy of Sciences of the United States of America. 85 (20): 7433–7. Bibcode:1988PNAS...85.7433N. doi:10.1073/pnas.85.20.7433. PMC 282205. PMID 2845406.
- ^ a b Chandra S, Kalaivani R, Kumar M, Srinivasan N, Sarkar DP (December 2017). Bassereau P (ed.). "Sendai virus recruits cellular villin to remodel actin cytoskeleton during fusion with hepatocytes". Molecular Biology of the Cell. 28 (26): 3801–3814. doi:10.1091/mbc.e17-06-0400. PMC 5739296. PMID 29074568.
- ^ a b Keskinen P, Nyqvist M, Sareneva T, Pirhonen J, Melén K, Julkunen I (October 1999). "Impaired antiviral response in human hepatoma cells". Virology. 263 (2): 364–75. doi:10.1006/viro.1999.9983. PMID 10544109.
- ^ a b c d e f g Shah NR, Sunderland A, Grdzelishvili VZ (June 2010). "Cell type mediated resistance of vesicular stomatitis virus and Sendai virus to ribavirin". PLOS ONE. 5 (6): e11265. Bibcode:2010PLoSO...511265S. doi:10.1371/journal.pone.0011265. PMC 2889835. PMID 20582319.
- ^ Sumpter R, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, et al. (March 2005). "Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I". Journal of Virology. 79 (5): 2689–99. doi:10.1128/JVI.79.5.2689-2699.2005. PMC 548482. PMID 15708988.
- ^ Lallemand C, Blanchard B, Palmieri M, Lebon P, May E, Tovey MG (January 2007). "Single-stranded RNA viruses inactivate the transcriptional activity of p53 but induce NOXA-dependent apoptosis via post-translational modifications of IRF-1, IRF-3 and CREB". Oncogene. 26 (3): 328–38. doi:10.1038/sj.onc.1209795. PMID 16832344. S2CID 25830890.
- ^ Buggele WA, Horvath CM (August 2013). "MicroRNA profiling of Sendai virus-infected A549 cells identifies miR-203 as an interferon-inducible regulator of IFIT1/ISG56". Journal of Virology. 87 (16): 9260–70. doi:10.1128/JVI.01064-13. PMC 3754065. PMID 23785202.
- ^ a b c d e f g Zainutdinov SS, Grazhdantseva AA, Kochetkov DV, Chumakov PM, Netesov SV, Matveeva OV, et al. (2017-10-01). "Change in Oncolytic Activity of Sendai Virus during Adaptation to Cell Cultures". Molecular Genetics, Microbiology and Virology. 32 (4): 212–217. doi:10.3103/S0891416817040115. S2CID 46958676.
- ^ Espada CE, Sari L, Cahill MP, Yang H, Phillips S, Martinez N, et al. (July 2023). "SAMHD1 impairs type I interferon induction through the MAVS, IKKε, and IRF7 signaling axis during viral infection". The Journal of Biological Chemistry. 299 (7): 104925. doi:10.1016/j.jbc.2023.104925. PMC 10404699. PMID 37328105.
- ^ a b c Belova AA, Sosnovtseva AO, Lipatova AV, Njushko KM, Volchenko NN, Belyakov MM, et al. (2017-01-01). "[Biomarkers of prostate cancer sensitivity to the Sendai virus]". Molekuliarnaia Biologiia. 51 (1): 94–103. doi:10.1134/S0026893317010046. PMID 28251971. S2CID 34514102.
- ^ a b Itoh M, Wang XL, Suzuki Y, Homma M (September 1992). "Mutation of the HANA protein of Sendai virus by passage in eggs". Virology. 190 (1): 356–64. doi:10.1016/0042-6822(92)91222-g. PMID 1326808.
- ^ a b Zainutdinov SS, Kochneva GV, Netesov SV, Chumakov PM, Matveeva OV (2019). "Directed evolution as a tool for the selection of oncolytic RNA viruses with desired phenotypes". Oncolytic Virotherapy. 8: 9–26. doi:10.2147/OV.S176523. PMC 6636189. PMID 31372363.
- ^ Sakaguchi T, Kiyotani K, Sakaki M, Fujii Y, Yoshida T (March 1994). "A field isolate of Sendai virus: its high virulence to mice and genetic divergence form prototype strains". Archives of Virology. 135 (1–2): 159–64. doi:10.1007/bf01309773. PMID 8198441. S2CID 12180965.
- ^ Itoh M, Isegawa Y, Hotta H, Homma M (December 1997). "Isolation of an avirulent mutant of Sendai virus with two amino acid mutations from a highly virulent field strain through adaptation to LLC-MK2 cells". The Journal of General Virology. 78 (12): 3207–15. doi:10.1099/0022-1317-78-12-3207. PMID 9400971.
- ^ Kiyotani K, Takao S, Sakaguchi T, Yoshida T (July 1990). "Immediate protection of mice from lethal wild-type Sendai virus (HVJ) infections by a temperature-sensitive mutant, HVJpi, possessing homologous interfering capacity". Virology. 177 (1): 65–74. doi:10.1016/0042-6822(90)90460-9. PMID 2162116.
- ^ a b Zhdanov VM, Bukrinskaya AG (1961). "Autoradiographic study of the penetration of Sendai virus into tissue culture cells. I. Preparation of Sendai virus labelled with radioactive isotopes". Problems of Virology. 6: 588–93. PMID 14040447.
- ^ Kiyotani K, Sakaguchi T, Fujii Y, Yoshida T (2001). "Attenuation of a field Sendai virus isolate through egg-passages is associated with an impediment of viral genome replication in mouse respiratory cells". Archives of Virology. 146 (5): 893–908. doi:10.1007/s007050170123. PMID 11448028. S2CID 21947750.
- ^ Kolakofsky D (August 1976). "Isolation and characterization of Sendai virus DI-RNAs". Cell. 8 (4): 547–555. doi:10.1016/0092-8674(76)90223-3. PMID 182384. S2CID 32399729.
- ^ Kolakofsky D (March 1979). "Studies on the generation and amplification of sendai virus defective-interfering genomes". Virology. 93 (2): 589–593. doi:10.1016/0042-6822(79)90263-0. PMID 222059.
- ^ Killip MJ, Young DF, Gatherer D, Ross CS, Short JA, Davison AJ, et al. (May 2013). "Deep sequencing analysis of defective genomes of parainfluenza virus 5 and their role in interferon induction". Journal of Virology. 87 (9): 4798–4807. doi:10.1128/JVI.03383-12. PMC 3624313. PMID 23449801.
- ^ Yoshida A, Kawabata R, Honda T, Sakai K, Ami Y, Sakaguchi T, et al. (March 2018). "A Single Amino Acid Substitution within the Paramyxovirus Sendai Virus Nucleoprotein Is a Critical Determinant for Production of Interferon-Beta-Inducing Copyback-Type Defective Interfering Genomes". Journal of Virology. 92 (5). doi:10.1128/JVI.02094-17. PMC 5809723. PMID 29237838.
- ^ Strahle L, Garcin D, Kolakofsky D (July 2006). "Sendai virus defective-interfering genomes and the activation of interferon-beta". Virology. 351 (1): 101–111. doi:10.1016/j.virol.2006.03.022. PMID 16631220.
- ^ Sánchez-Aparicio MT, Garcin D, Rice CM, Kolakofsky D, García-Sastre A, Baum A (June 2017). "Loss of Sendai virus C protein leads to accumulation of RIG-I immunostimulatory defective interfering RNA". The Journal of General Virology. 98 (6): 1282–1293. doi:10.1099/jgv.0.000815. PMC 5962894. PMID 28631605.
- ^ Tatsumoto N, Arditi M, Yamashita M (September 2018). "Sendai Virus Propagation Using Chicken Eggs". Bio-Protocol. 8 (18). doi:10.21769/BioProtoc.3009. PMC 6200407. PMID 30370318.
- ^ Pappas C, Matsuoka Y, Swayne DE, Donis RO (November 2007). "Development and evaluation of an Influenza virus subtype H7N2 vaccine candidate for pandemic preparedness". Clinical and Vaccine Immunology. 14 (11): 1425–1432. doi:10.1128/CVI.00174-07. PMC 2168170. PMID 17913860.
- ^ Racaniello V (13 July 2009). "Measurement of viruses by end-point dilution assay". Virology Blog. 27: 493–497.
- ^ Tatsumoto N, Miyauchi T, Arditi M, Yamashita M (November 2018). "Quantification of Infectious Sendai Virus Using Plaque Assay". Bio-Protocol. 8 (21). doi:10.21769/BioProtoc.3068. PMC 6289198. PMID 30547053.
- ^ Killian ML (2008). "Hemagglutination Assay for the Avian Influenza Virus". Avian Influenza Virus. Methods in Molecular Biology. Vol. 436. Totowa, NJ: Humana Press. pp. 47–52. doi:10.1007/978-1-59745-279-3_7. ISBN 978-1-58829-939-0. PMID 18370040.
