Virus quantification
Virus quantification is counting or calculating the number of virus particles (virions) in a sample to determine the virus concentration. It is used in both research and development (R&D) in academic and commercial laboratories as well as in production situations where the quantity of virus at various steps is an important variable that must be monitored. For example, the production of virus-based vaccines, recombinant proteins using viral vectors, and viral antigens all require virus quantification to continually monitor and/or modify the process in order to optimize product quality and production yields and to respond to ever changing demands and applications. Other examples of specific instances where viruses need to be quantified include clone screening, multiplicity of infection (MOI) optimization, and adaptation of methods to cell culture.
There are many ways to categorize virus quantification methods. Here, the methods are grouped according to what is being measured and in what biological context. For example, cell-based assays typically measure infectious units (active virus). Other methods may measure the concentration of viral proteins, DNA, RNA, or molecular particles, but not necessarily measure infectivity. Each method has its own advantages and disadvantages, which often determines which method is used for specific applications.[1]
Cell-based assays
[edit]Plaque assay
[edit]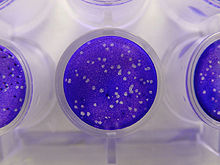
Plaque-based assays are a commonly used method to determine virus concentration in terms of infectious dose. Plaque assays determine the number of plaque forming units (PFU) in a virus sample, which is one measure of virus quantity. This assay is based on a microbiological method conducted in petri dishes or multi-well cell culture plates. Specifically, a confluent monolayer of host cells is infected by applying a sample containing the virus at varying dilutions and then covered with a semi-solid medium, such as agar or carboxymethyl cellulose, to prevent the virus infection from spreading indiscriminately, as would occur in a liquid medium. A viral plaque is formed after a virus infects a cell within the fixed cell monolayer.[2] The virus-infected cell will lyse and spread the infection to adjacent cells, where the infection-to-lysis cycle is repeated. This will create an area of infected, lysed cells (viral plaque) surrounded by uninfected, intact cells. The plaque can be seen with an optical microscope or visually using cell staining techniques (e.g., staining with a crystal violet solution to visualize intact vs. lysed cells).[3] Plaque formation can take 3–14 days, depending on the virus being analyzed. Plaques are generally counted manually, and the plaque count, in combination with the dilution factor of the infection solution (the sample initially applied to the cells), is used to calculate the number of plaque forming units per sample unit volume (PFU/mL). The PFU/mL number represents the concentration of infectious virus particles within the sample and is based on the assumption that each plaque formed is representative of an initial infection by one infectious virus particle.[4][5]
Focus forming assay (FFA)
[edit]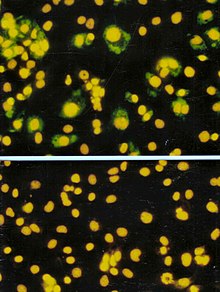
The focus forming assay (FFA) is a variation of the plaque assay, but instead of depending on cell lysis in order to detect plaque formation, the FFA employs immunostaining techniques using fluorescently labeled antibodies specific for a viral antigen to detect infected host cells and infectious virus particles before an actual plaque is formed. The FFA is particularly useful for quantifying classes of viruses that do not lyse the cell membranes, as these viruses would not be amenable to the plaque assay. Like the plaque assay, host cell monolayers are infected with various dilutions of the virus sample and allowed to incubate for a relatively brief incubation period (e.g., 24–72 hours) under a semisolid overlay medium that restricts the spread of infectious virus, creating localized clusters (foci) of infected cells. Plates are subsequently probed with fluorescently labeled antibodies against a viral antigen, and fluorescence microscopy is used to count and quantify the number of foci. The FFA method typically yields results in less time than plaque assays or fifty-percent-tissue-culture-infective-dose (TCID50) assays (see below), but it can be more expensive in terms of required reagents and equipment. Assay completion time is also dependent on the size of area that the user is counting. A larger area will require more time but can provide a more accurate representation of the sample. Results of the FFA are expressed as focus forming units per milliliter, or FFU/mL.[6]
TCID50 endpoint dilution assay
[edit]The TCID50 (50% tissue culture infectious dose) assay is the measure of infectious virus titer. This endpoint dilution assay quantifies the amount of virus required to kill 50% of infected hosts or to produce a cytopathic effect in 50% of inoculated tissue culture cells. This assay may be more common in clinical research applications where the lethal dose of virus must be determined or if the virus does not form plaques.[citation needed] When used in the context of tissue culture, host cells are plated and serial dilutions of the virus are added. After incubation, the percentage of cell death (i.e. infected cells) is manually observed and recorded for each virus dilution, and results are used to mathematically calculate a TCID50 result.[6][7] Due to distinct differences in assay methods and principles, TCID50 and pfu/mL or other infectivity assay results are not equivalent. This method can take up to a week due to cell infectivity time.[8]
Two methods commonly used to calculate TCID50 (can also be used to calculate other types of 50% endpoint such EC50, IC50, and LD50) are:
- Spearman–Kärber[9]
- Reed–Muench method
The theoretical relationship between TCID50 and PFU is approximately 0.69 PFU = 1 TCID50 based on the Poisson distribution,[10] a probability distribution which describes how many random events (virus particles) occurring at a known average rate (virus titer) are likely to occur in a fixed space (the amount of virus medium in a well). However, it must be emphasized that in practice, this relationship may not hold even for the same virus + cell combination, as the two types of assay are set up differently and virus infectivity is very sensitive to various factors such as cell age, overlay media, etc. But the following reference defines the relationship differently:
From ATTC: "Assuming that the same cell system is used, that the virus forms plaques on those cells, and that no procedures are added which would inhibit plaque formation, 1 mL of virus stock would be expected to have about half of the number of plaque forming units (PFUs) as TCID50. This is only an estimate but is based on the rationale that the limiting dilution which would infect 50% of the cell layers challenged would often be expected to initially produce a single plaque in the cell layers which become infected. In some instances, two or more plaques might by chance form, and thus the actual number of PFUs should be determined experimentally.
"Mathematically, the expected PFUs would be somewhat greater than one-half the TCID50, since the negative tubes in the TCID50 represent zero plaque forming units and the positive tubes each represent one or more plaque forming units. A more precise estimate is obtained by applying the Poisson distribution. Where is the proportion of negative tubes and m is the mean number of infectious units per volume (PFU/ml), . For any titer expressed as a TCID50, . Thus and which is ~ 0.7.
"Therefore, one could multiply the TCID50 titer (per ml) by 0.7 to predict the mean number of PFU/ml. When actually applying such calculations, remember the calculated mean will only be valid if the changes in protocol required to visualize plaques do not alter the expression of infectious virus as compared with expression under conditions employed for TCID50.
"Thus as a working estimate, one can assume material with a TCID50 of 1 × 105 TCID50/mL will produce 0.7 × 105 PFUs/mL." [11]
Protein and antibody-based assays
[edit]There are several variations of protein- and antibody-based virus quantification assays. In general, these methods quantify either the amount of all protein or the amount of a specific virus protein in the sample rather than the number of infected cells or virus particles. Quantification commonly relies on colorimetric or fluorescence detection. Some assay variations quantify proteins directly in a sample, while other variations require host cell infection and incubation to allow virus growth prior to quantification. The variation used depends primarily on the amount of protein (i.e. viral protein) in the initial sample and the sensitivity of the assay itself. If incubation and virus growth are required, cell and/or virus lysis/digestion are often conducted prior to analysis. Most protein-based methods are relatively fast and sensitive[citation needed] but require quality standards for accurate calibration, and quantify protein, not actual virus particle concentrations. Below are specific examples of widely used protein-based assays.
Hemagglutination assay
[edit]The hemagglutination assay (HA) is a common non-fluorescence protein quantification assay specific for influenza. It relies on the fact that hemagglutinin, a surface protein of influenza viruses, agglutinates red blood cells (i.e. causes red blood cells to clump together). In this assay, dilutions of an influenza sample are incubated with a 1% erythrocyte solution for one hour and the virus dilution at which agglutination first occurs is visually determined. The assay produces a result of hemagglutination units (HAU), with typical PFU to HAU ratios in the 106 range.[12][13][14] This assay takes ~1–2 hours to complete.
The hemagglutination inhibition assay is a common variation of the HA assay used to measure flu-specific antibody levels in blood serum. In this variation, serum antibodies to the influenza virus will interfere with the virus attachment to red blood cells. Therefore, hemagglutination is inhibited when antibodies are present at a sufficient concentration.[15]
Bicinchoninic acid (BCA) assay
[edit]The bicinchoninic acid assay (BCA; a.k.a. Smith assay) is based on a simple colorimetric measurement and is a commonly used protein quantification assay.[16] BCA is similar to the Lowry or Bradford protein assays. The BCA assay reagent was first developed and made commercially by Pierce Chemical Company (now owned by Thermo Fisher Scientific) which held the patent until 2006.[17][18]
In the BCA assay, a protein's peptide bonds quantitatively reduce Cu2+ to Cu1+, which produces a light blue color. BCA chelates Cu1+ at a 2:1 ratio resulting in a more intensely colored species that absorbs at562 nm. Absorbance of a sample at 562 nm is used to determine the bulk protein concentration in the sample. Assay results are compared with known standard curves after analysis with a spectrophotometer or plate reader.[19] Total assay time is 30 minutes to one hour. While this assay is ubiquitous and fast, it lacks specificity to viral proteins since it counts all protein in the sample. Thus the virus preparation to be quantified must contain very low levels of host cell proteins.
Enzyme-linked immunosorbent assay (ELISA)
[edit]
Enzyme-linked immunosorbent assay (ELISA) is an antibody-based assay that utilizes an antigen-specific antibody chemically linked to an enzyme (or bound to a second antibody linked to an enzyme) to detect the presence of an unknown amount of the antigen (e.g., viral protein) in a sample. The antibody-antigen binding event is detected and/or quantified through the enzyme's ability to convert a substrate reagent to produce a detectable signal that can then be used to calculate the concentration of the target antigen in the sample.[20] Horseradish peroxidase (HRP) is a common enzyme utilized in ELISA schemes due to its ability to amplify signal and increase assay sensitivity.
There are many variations, or types of ELISA assays but they can generally be classified as either indirect, competitive, sandwich or reverse.[21]
Single radial immunodiffusion (SRID) assay
[edit]Single radial immunodiffusion assay (SRID), also known as the Mancini method, is a protein assay that detects the amount of specific viral antigen by immunodiffusion in a semi-solid medium (e.g. agar). The medium contains antiserum specific to the antigen of interest and the antigen is placed in the center of the disc. As the antigen diffuses into the medium it creates a precipitate ring that grows until equilibrium is reached. Assay time can range from 10 hours to days depending on equilibration time of the antigen and antibody. The zone diameter from the ring is linearly related to the log of protein concentration and is compared to zone diameters for known protein standards for quantification.[22]
DNA and RNA assays
[edit]Quantitative polymerase chain reaction (qPCR)
[edit]Quantitative PCR utilizes polymerase chain reaction chemistry to amplify viral DNA or RNA to produce high enough concentrations for detection and quantification by fluorescence. In general, quantification by qPCR relies on serial dilutions of standards of known concentration being analyzed in parallel with the unknown samples for calibration and reference. Quantitative detection can be achieved using a wide variety of fluorescence detection strategies, including sequence specific probes or non-specific fluorescent dyes such as SYBR Green.[23] Sequence-specific probes, such as TaqMan Molecular Beacons, or Scorpion, bind only to the DNA of the appropriate sequence produced during the reaction. SYBR Green dye binds to all double-stranded DNA[24] produced during the reaction.
While SYBR Green is easy to use, its lack of specificity and lower sensitivity lead most labs to use probe-based qPCR detection schemes.[citation needed] There are many variations of qPCR including the comparative threshold method, which allows relative quantification through comparison of Ct values (PCR cycles that show statistically significant increases in the product) from multiple samples that include an internal standard.[25]
PCR amplifies all target nucleic acid, including ones originating from intact infectious viral particles, from defective viral particles as well as free nucleic acid in solution. Because of this, qPCR results (expressed in terms of genome copies/mL) are likely to be higher in quantity than TEM results. For viral quantification, the ratio of whole virions to copies of nucleic acid is seldom one to one. This is because during viral replication, the nucleic acid and viral proteins are not always produced in 1:1 ratio and viral assembly process results in complete virions as well as empty capsids and/or excess free viral genomes. In the example of foot-and-mouth disease virus, the ratio of whole virions to RNA copies within an actively replicating host cell is approximately 1:1000.[26] Advantages of titration by qPCR include quick turnaround time (1–4 hours) and sensitivity (can detect much lower concentration of viruses than other methods).
Particle Assays
[edit]Tunable resistive pulse sensing (TRPS)
[edit]Tunable resistive pulse sensing (TRPS) is a method that allows high-throughput single particle measurements of individual virus particles, as they are driven through a size-tunable nanopore, one at a time.[27] The technique has the advantage of simultaneously determining the size and concentration, of virus particles in solution with high resolution. This can be used in assessing sample stability and the contribution of aggregates, as well as total viral particle concentration (vp/mL).[28]
TRPS-based measurement occurs in an ionic buffer, and no pre-staining of samples is required prior to analysis, thus the technique is more rapid than those which require pre-treatment with fluorescent dyes, with a total preparation and measurement time of less than 10 minutes per sample.[citation needed] TRPS-bases virus analysis is commercially available through qViro-X systems, which have the ability to be decontaminated chemically by autoclaving after measurement has occurred.
Single Virus Inductively Coupled Plasma Mass Spectroscopy (SV ICP-MS)
[edit]This technique is similar to Single Particle Inductively Coupled Plasma Mass Spectroscopy (SP ICP-MS) discovered by Degueldre and Favarger (2003)[29] and adapted later for other nanoparticles (e.g. gold colloids, see Degueldre et al. (2006)).[30] The SP ICP-MS was adapted for the analysis of Single Virus Inductively Coupled Plasma Mass Spectroscopy (SV ICPMS) in a comprehensive study i.e. Degueldre (2021).[31] This study suggests to adapting this method for single viruses (SV) identification and counting. With high resolution multi-channel sector field (MC SF) ICP-MS records in SV detection mode, the counting of master and key ions can allow analysis and identification of single viruses. The counting of 2-500 virial units can be performed in 20 s. Analyses are proposed to be carried out in Ar torch for master ions: 12C+, 13C+, 14N+, 15N+, and key ions 31P+, 32S+, 33S+ and 34S+. All interferences are discussed in detail. The use of high resolution MC ICP-MS is recommended while options with anaerobic/aerobic atmospheres are explored to upgrade the analysis when using quadrupole ICP-MS. Application for two virus types (SARS-COV2 and bacteriophage T5) is investigated using time scan and fixed mass analysis for the selected virus ions allowing characterisation of the species using the N/C, P/C and S/C molar ratio's and quantification of their number concentration.
Other assays
[edit]Flow cytometry
[edit]While most flow cytometers do not have sufficient sensitivity,[citation needed] there are a few commercially available flow cytometers that can be used for virus quantification. A virus counter quantifies the number of intact virus particles in a sample using fluorescence to detect colocalized proteins and nucleic acids. Samples are stained with two dyes, one specific for proteins and one specific for nucleic acids, and analyzed as they flow through a laser beam. The quantity of particles producing simultaneous events on each of the two distinct fluorescence channels is determined, along with the measured sample flow rate, to calculate a concentration of virus particles (vp/mL).[32] The results are generally similar in absolute quantity to a TEM result. The assay has a linear working range of 105–109 vp/mL and an analysis time of ~10 min with a short sample preparation time.[citation needed]
Transmission electron microscopy (TEM)
[edit]TEM is a specialized type of microscopy that utilizes a beam of electrons focused with a magnetic field to image a sample. TEM provides imaging with 1000x greater spatial resolution than a light microscope (resolution down to 0.2 nm).[33] An ultrathin, negatively stained sample is required. Sample preparations involve depositing specimens onto a coated TEM grid and negative staining with an electron-opaque liquid.[34] Tissue embedded samples can also be examined if thinly sectioned. Sample preparations vary depending on protocol and user but generally require hours to complete. TEM images can show individual virus particles and quantitative image analysis can be used to determine virus concentrations. These high resolution images also provide particle morphology information that most other methods cannot. Quantitative TEM results will often be greater than results from other assays[citation needed] as all particles, regardless of infectivity, are quantified in the reported virus-like particles per mL (vlp/mL) result. Quantitative TEM generally works well for virus concentrations greater than 106 particles/mL. Because of high instrument cost and the amount of space and support facilities needed, TEM equipment is only available in a few laboratories.
See also
[edit]References
[edit]- ^ Sciences, Noble Life. "Viral Quantification Methods". content.noblelifesci.com. Retrieved 2023-08-07.
- ^ Kaufmann, S.H.; Kabelitz, D. (2002). Methods in Microbiology Vol.32:Immunology of Infection. Academic Press. ISBN 0-12-521532-0.
- ^ Baer, Alan; Kehn-Hall, Kylene (November 4, 2014). "Viral Concentration Determination Through Plaque Assays: Using Traditional and Novel Overlay Systems". Journal of Visualized Experiments (93): e52065. doi:10.3791/52065. PMC 4255882. PMID 25407402.
- ^ Martin, S.J. (1978). The Biochemistry of Viruses. Cambridge University Press. ISBN 0-12-402033-X.
- ^ Yakimovich, Artur; Andriasyan, Vardan; Witte, Robert; Wang, I.-Hsuan; Prasad, Vibhu; Suomalainen, Maarit; Greber, Urs F. (2015-09-28). "Plaque2.0—A High-Throughput Analysis Framework to Score Virus-Cell Transmission and Clonal Cell Expansion". PLOS ONE. 10 (9): e0138760. Bibcode:2015PLoSO..1038760Y. doi:10.1371/journal.pone.0138760. ISSN 1932-6203. PMC 4587671. PMID 26413745.
- ^ a b Flint, S.J.; Enquist, W.; Racaniello, V.R.; Skalka, A.M. (2009). "Virological Methods". Principles of Virology. ASM Press. ISBN 978-1-55581-443-4.
- ^ Lindenbach, Brett D. "Reed & Muench Calculator".
- ^ "Archived copy" (PDF). Archived from the original (PDF) on 2011-09-27. Retrieved 2010-02-26.
{{cite web}}: CS1 maint: archived copy as title (link) - ^ Kärber, G. (1931). "Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche". Naunyn-Schmiedebergs Archiv für Experimentelle Pathologie und Pharmakologie. 162 (4). Springer-Verlag: 480–483. doi:10.1007/BF01863914. S2CID 46017573.
- ^ thneedle (16 April 2002). "TCID50 and plaque forming unit (PFU)". Retrieved 29 May 2014.
- ^ "Virology Culture Guide". www.atcc.org. Retrieved 2023-08-09.
- ^ Killian, M.L. (2008). "Hemagglutination Assay for the Avian Influenza Virus". In Spackman, Erica (ed.). Avian Influenza Virus. Vol. 436. Humana Press. pp. 47–52. doi:10.1007/978-1-59745-279-3_7. ISBN 978-1-58829-939-0. PMID 18370040.
- ^ Rimmelzwaan, G.F.; Baars, M.; Claas, E.C.J.; Osterhaus, A.D.M.E. (1998). "Comparison of RNA Hybridization, Hemaaglutination Assay, Titration of Infectious Virus and Immunofluorescence as Methods for Monitoring Influenza Virus Replication In Vitro". Journal of Virological Methods. 74 (1): 57–66. doi:10.1016/S0166-0934(98)00071-8. PMID 9763129.
- ^ Kato, A.; Kiyotani, K.; Sakai, Y.; Yoshida, T.; Nagai, Y. (1997). "The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis". The EMBO Journal. 16 (3): 578–587. doi:10.1093/emboj/16.3.578. PMC 1169661. PMID 9034340.
- ^ "Influenza hemagglutination inhibition assay". 27 May 2009.
- ^ "Bicinchoninic Acid Assay - an overview | ScienceDirect Topics". www.sciencedirect.com. Retrieved 2023-08-07.
- ^ Smith, P. K.; Krohn, R. I.; Hermanson, G. T.; Mallia, A. K.; Gartner, F. H.; Provenzano, M. D.; Fujimoto, E. K.; Goeke, N. M.; Olson, B. J.; Klenk, D. C. (1985-10-01). "Measurement of protein using bicinchoninic acid". Analytical Biochemistry. 150 (1): 76–85. doi:10.1016/0003-2697(85)90442-7. ISSN 0003-2697. PMID 3843705.
- ^ US Patent and Trademark Office. "Measurement of protein using bicinchoninic acid". Google Patents. Retrieved 2023-08-07.
- ^ "Pierce Protein Biology".
- ^ Kemeny, D.M.; Challacombe, S.J. (1988). ELISA and Other Solid Phase Immunoassays: Theoretical and Practical Aspects. John Wiley and Sons. ISBN 0-471-90982-3.
- ^ Kuby, J.; Kindt, T.J.; Goldsby, R.A.; Osborne, B.A. (2007). Kuby Immunology 6th edition. W.H. Freeman and Company. ISBN 978-1-4292-0211-4.
- ^ Rodda, S.J.; Gallichio, H.A.; Hampson, A.W (1981). "The Single Radial Immunodiffusion Assay Highlights Small Antigenic Differences Among Influenza Virus Hemagglutinins". Journal of Clinical Microbiology. 14 (5): 479–482. doi:10.1128/JCM.14.5.479-482.1981. PMC 273972. PMID 6171580.
- ^ "PCR/Real-Time PCR Protocols".
- ^ "Applied Biosystems - US" (PDF).
- ^ O'Leary, J.J.; Sheils, O.; Martin, C.; Crowley, A. (2003). "Taqman Technology and Real-Time Polymerase Chain Reaction". In Crocker, J.; Murray, P.G. (eds.). Molecular Biology in Cellular Pathology. John Wiley and Sons. pp. 251–268. ISBN 978-0-470-84475-5.
- ^ Callahan JD, et al., Use of a portable real-time reverse transcriptase-polymerase chain reaction assay for rapid detection of foot-and-mouth disease virus. J Am Vet Med Assoc. 2002 Jun 1;220(11):1636-42.
- ^ Stephen J. Sowerby, Murray F. Broom, George B. Petersen. "Dynamically resizable nanometre-scale apertures for molecular sensing" Sensors and Actuators B: Chemical Volume 123, Issue 1 (2007), pages 325-330
- ^ G. Seth Roberts, Sam Yu, Qinglu Zeng, Leslie C.L. Chan, Will Anderson, Aaron H. Colby, Mark W. Grinstaff, Steven Reid, Robert Vogel. “Tunable Pores for Measuring Concentrations of Synthetic and Biological Nanoparticle Dispersions” Biosensors and Bioelectronics, 31 pp. 17-25, (2012).
- ^ C. Degueldre, P.-Y. Favarger, Colloid analysis by single particle inductively coupled plasma-mass spectroscopy: a feasibility study, Colloids and Surfaces A, 217 (2003) 137-142.
- ^ C Degueldre, P.-Y Favarger, S. Wold, Gold colloid analysis by single particle inductively coupled plasma-mass spectrometry in a single particle mode, Anal Chim Acta, 555 (2006) 263-268.
- ^ C. Degueldre, Single virus inductively coupled plasma mass spectroscopy analysis: a comprehensive study. Talanta, Vol 228 Feb (2021) 122211.
- ^ Stoffel, C.L.; Finch, R.; Christensen, K.; Edwards, D.; Rowlen, K.L. (2005). "Rapid Determination of Baculovirus Titer by a Dual Channel Virus Counter". American Biotechnology Laboratory. 37 (22): 24–25.
- ^ Sherman, I. "Resolution of an Electron Microscope". The Physics Factbook. Retrieved February 25, 2010.
- ^ Steffens, W.L. (1998). "Use of Transmission Electron Microscopy for Viral Diagnosis in Psittacine Birds". Proceedings of International Virtual Conferences in Veterinary Medicine: Diseases of Psittacine Birds. Athens, Georgia.







