RNA silencing suppressor p19
| RNA silencing suppressor p19 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 The protein dimer formed by two p19 molecules. Each monomer is colored from N-terminus (blue) to C-terminus (red) to illustrate the end-to-end orientation of the dimer. The dotted gray line in the center highlights the dimer interface. From PDB: 1R9F.[1] | |||||||||
| Identifiers | |||||||||
| Symbol | Tombus_p19 | ||||||||
| Pfam | PF03220 | ||||||||
| InterPro | IPR004905 | ||||||||
| |||||||||
RNA silencing suppressor p19 (also known as Tombusvirus P19 core protein and 19 kDa symptom severity modulator) is a protein expressed from the ORF4 gene in the genome of tombusviruses. These viruses are positive-sense single-stranded RNA viruses that infect plant cells, in which RNA silencing forms a widespread and robust antiviral defense system. The p19 protein serves as a counter-defense strategy, specifically binding the 19- to 21-nucleotide double-stranded RNAs that function as small interfering RNA (siRNA) in the RNA silencing system. By sequestering siRNA, p19 suppresses RNA silencing and promotes viral proliferation.[1][2][3] The p19 protein is considered a significant virulence factor[4] and a component of an evolutionary arms race between plants and their pathogens.[5]
Structure
[edit]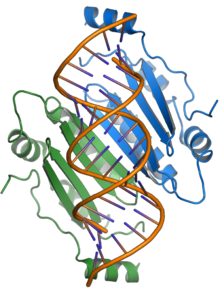
The p19 protein received its name from its size, being approximately 19 kilodaltons. It forms a functional homodimer. The crystal structures are available of p19 proteins from the tomato bushy stunt virus[1] and Carnation Italian ringspot virus;[2] the protein consists of a novel protein fold and exemplifies a previously unknown mechanism for binding RNA, using a binding surface formed by a beta sheet and flanked by alpha helices to interact with double-stranded RNAs of around 21 nucleotides in length in a non-sequence-specific manner.[1][2][7]
Function
[edit]The p19 protein binds to double-stranded RNAs that function as short interfering RNA (siRNA) and is specialized for the 21-nucleotide product of the enzyme DCL4 (a member of a family of plant enzymes with homology to Dicer).[7] By binding to siRNA, p19 sequesters these species and prevents them from interacting with the RNA-induced silencing complex (RISC), a protein complex that mediates the antiviral RNA silencing mechanism in the cell.
The p19 protein is also capable of binding to microRNA molecules that are endogenous to the host cell, as well as the siRNAs that are ultimately derived from the virus's own genome. Notably, an exception to this pattern is p19's inefficiency in interacting with the microRNA miR-168, a regulatory non-coding RNA that represses expression of argonaute-1 (AGO1). The AGO1 protein is required for RNA silencing, thus selectively sparing its repressor from p19's general sequestration of miRNA has the effect of reducing cellular AGO1 levels and is an additional mechanism by which p19 inhibits silencing.[5][8] The two mechanisms are independent of one another and can be selectively abrogated by mutations.[9]
Evolution
[edit]The gene encoding the p19 protein is an example of an overprinted gene, a genomic arrangement common in viruses in which multiple genes are encoded by the same portion of the genome read in alternate reading frames.[10][11] The open reading frame ORF4, which encodes p19, is completely contained within the open reading frame of another gene, which is designated ORF3 and encodes the movement protein p22. Both genes, and their relative positions, are conserved within the tombusvirus family.[4][11] P19 is thought to have originated de novo in this lineage.[11][12]
Sequestration of dsRNA is a common viral counter-defense strategy against RNA silencing, evolved in a form of evolutionary arms race between virus and host.[5] The p19 protein is not unique in this role; in an example of convergent evolution, this strategy appears to have evolved at least three times in distinct viral lineages using proteins with distinct structures and physical means of binding RNA.[3][13][14]
History
[edit]The tomato bushy stunt virus, which is the type species of the tombusvirus family, is a long-standing model system for the study of plant viruses. The open reading frame encoding p19 was originally discovered in the late 1980s when the virus's genome was sequenced; it was subsequently demonstrated that the predicted protein was indeed expressed from the gene, although its role in promoting virulence and infectivity was initially underappreciated. Following the elucidation of its role as a suppressor of RNA silencing, p19 has also been used as a tool in molecular biology research on RNA silencing, RNA interference, and related processes.[4][6]
References
[edit]- ^ a b c d e Ye K, Malinina L, Patel DJ (December 2003). "Recognition of small interfering RNA by a viral suppressor of RNA silencing". Nature. 426 (6968): 874–8. Bibcode:2003Natur.426..874Y. doi:10.1038/nature02213. PMC 4694583. PMID 14661029.
- ^ a b c Vargason JM, Szittya G, Burgyán J, Hall TM (December 2003). "Size selective recognition of siRNA by an RNA silencing suppressor". Cell. 115 (7): 799–811. doi:10.1016/S0092-8674(03)00984-X. PMID 14697199. S2CID 12993441.
- ^ a b Lakatos L, Szittya G, Silhavy D, Burgyán J (February 2004). "Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses". The EMBO Journal. 23 (4): 876–84. doi:10.1038/sj.emboj.7600096. PMC 381004. PMID 14976549.
- ^ a b c Scholthof HB (May 2006). "The Tombusvirus-encoded P19: from irrelevance to elegance". Nature Reviews. Microbiology. 4 (5): 405–11. doi:10.1038/nrmicro1395. PMID 16518419. S2CID 30361458.
- ^ a b c Pumplin N, Voinnet O (November 2013). "RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence". Nature Reviews. Microbiology. 11 (11): 745–60. doi:10.1038/nrmicro3120. PMID 24129510. S2CID 205498691.
- ^ a b Danielson DC, Pezacki JP (April 2013). "Studying the RNA silencing pathway with the p19 protein". FEBS Letters. 587 (8): 1198–205. doi:10.1016/j.febslet.2013.01.036. PMID 23376479.
- ^ a b Ding SW, Voinnet O (August 2007). "Antiviral immunity directed by small RNAs". Cell. 130 (3): 413–26. doi:10.1016/j.cell.2007.07.039. PMC 2703654. PMID 17693253.
- ^ Várallyay E, Válóczi A, Agyi A, Burgyán J, Havelda Z (October 2010). "Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation". The EMBO Journal. 29 (20): 3507–19. doi:10.1038/emboj.2010.215. PMC 2964164. PMID 20823831.
- ^ Várallyay É, Oláh E, Havelda Z (January 2014). "Independent parallel functions of p19 plant viral suppressor of RNA silencing required for effective suppressor activity". Nucleic Acids Research. 42 (1): 599–608. doi:10.1093/nar/gkt846. PMC 3874164. PMID 24062160.
- ^ Scholthof HB, Scholthof KB, Kikkert M, Jackson AO (November 1995). "Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion". Virology. 213 (2): 425–38. doi:10.1006/viro.1995.0015. PMID 7491767.
- ^ a b c Rancurel C, Khosravi M, Dunker AK, Romero PR, Karlin D (October 2009). "Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation". Journal of Virology. 83 (20): 10719–36. doi:10.1128/JVI.00595-09. PMC 2753099. PMID 19640978.
- ^ Sabath N, Wagner A, Karlin D (December 2012). "Evolution of viral proteins originated de novo by overprinting". Molecular Biology and Evolution. 29 (12): 3767–80. doi:10.1093/molbev/mss179. PMC 3494269. PMID 22821011.
- ^ Lakatos L, Csorba T, Pantaleo V, Chapman EJ, Carrington JC, Liu YP, Dolja VV, Calvino LF, López-Moya JJ, Burgyán J (June 2006). "Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors". The EMBO Journal. 25 (12): 2768–80. doi:10.1038/sj.emboj.7601164. PMC 1500863. PMID 16724105.
- ^ Voinnet O (March 2005). "Induction and suppression of RNA silencing: insights from viral infections". Nature Reviews. Genetics. 6 (3): 206–20. doi:10.1038/nrg1555. PMID 15703763. S2CID 26351712.
