Phomoxanthone A
 | |
| Names | |
|---|---|
| Preferred IUPAC name
rel-(5R,5′R,6R,6′R,10aR,10′aR)-10a,10′a-Bis[(acetyloxy)methyl]-1,1′,8,8′-tetrahydroxy-6,6′-dimethyl-9,9′-dioxo-5,5′,7,7′,9,9′,10a,10′a-octahydro-6H,6′H-[4,4′-bixanthene]-5,5′-diyl diacetate | |
| Other names
PXA
| |
| Identifiers | |
3D model (JSmol)
|
|
| ChEBI | |
| ChEMBL | |
| ChemSpider | |
PubChem CID
|
|
CompTox Dashboard (EPA)
|
|
| |
| |
| Properties | |
| C38H38O16 | |
| Molar mass | 750.70 g/mol |
| Appearance | yellow solid |
| Density | ~1.53 g/cm3 |
| not soluble | |
| Solubility in DMSO | good, but unstable[1] |
| Solubility in EtOH | moderate[1] |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
The mycotoxin phomoxanthone A, or PXA for short, is a toxic natural product that affects the mitochondria. It is the most toxic and the best studied of the naturally occurring phomoxanthones. PXA has recently been shown to induce rapid, non-canonical mitochondrial fission by causing the mitochondrial matrix to fragment while the outer mitochondrial membrane can remain intact. This process was shown to be independent from the mitochondrial fission and fusion regulators DRP1 and OPA1.[1]
Properties and structure
[edit]
The phomoxanthones are named after the fungus Phomopsis, from which they were first isolated, and after their xanthonoid structure, which means they have structures similar to the compound xanthone (pictured on the left). Chemically, the phomoxanthones are dimers of two tetrahydroxanthones, meaning that they consist of two subunits of xanthonoids that have four hydroxy groups each. The two subunits of the phomoxanthones are covalently linked to each other. PXA itself is a homodimer, meaning that it consists of two identical subunits. Both of these subunits are diacetylated tetrahydroxanthones, so two of their hydroxy groups have been replaced by acetyl groups. The position of the link between the two dimer subunits is the only structural difference between PXA and its less toxic isomers phomoxanthone B (PXB) and dicerandrol C: In PXA, the two xanthonoid monomers are symmetrically linked at the position C-4,4’, while in PXB, they are asymmetrically linked at C-2,4’, and in dicerandrol C, they are symmetrically linked at C-2,2’. Otherwise, these three compounds are structurally identical.[2][3] The phomoxanthones are structurally closely related to the secalonic acids, another class of dimeric tetrahydroxanthone mycotoxins, with which they share several properties. Notably, both the phomoxanthones and the secalonic acids are unstable when dissolved in polar solvents such as DMSO, with the covalent bond between the two monomers shifting between 2,2′-, 2,4′-, and 4,4′-linkage.[4] The two phomoxanthones PXA and PXB can thus slowly isomerise into each other as well as into the essentially non-toxic dicerandrol C, resulting in a loss of activity of PXA over time when dissolved in a polar solvent.[1]
Occurrence
[edit]As natural products, PXA and other phomoxanthones occur as secondary metabolites in fungi of the eponymous genus Phomopsis, most notably in the species Phomopsis longicolla.[2][3] This fungus is an endophyte of the mangrove plant Sonneratia caseolaris.[5][3] However, it has also been identified as a pathogen in other plants, such as the soybean plant in which it causes a disease called Phomopsis seed decay (PSD).[6][7]
Preparation
[edit]Both PXA and PXB were discovered in 2001, and their preparation by isolation from Phomopsis fungal cultures was described in the corresponding publication.[2] Briefly, a MeOH extract of a Phomopsis culture is mixed with H2O and washed with hexane. The aqueous phase is then dried and the residue is dissolved in EtOAc, washed with H2O, concentrated and repeatedly purified by size-exclusion chromatography. The resulting mixture of PXA and PXB is separated by HPLC. A modified method, in which the initial extraction is done with EtOAc instead of MeOH and the drying step is skipped, was described in 2013.[3]
Uses
[edit]Phomoxanthone A was first identified in a screening for antimalarial compounds.[2] It showed strong antibiotic activity against a multidrug-resistant strain of the main causative agent of malaria, the protozoan parasite Plasmodium falciparum. The same study also reported antibiotic activity of PXA against Mycobacterium tuberculosis and against three animal cell lines, two of which were derived from human cancer cells.[2] These findings not only showed that PXA has antibiotic activity against very diverse organisms, but they also sparked further studies that investigated PXA as a potential antibiotic or anti-cancer drug. A later study also reported antibiotic activity for PXA against the alga Chlorella fusca, the fungus Ustilago violacea, and the bacterium Bacillus megaterium.[8] This broad range of activity disqualified it as a specific antibiotic that could be used in the treatment of infectious diseases, however the hope that it could be used as an anti-cancer drug remained. Preliminary results from a study in human cancer cells and non-cancer cells suggested that PXA might be more toxic to the former than to the latter, although results from in vivo studies have not yet been presented.[3][9]
Aside from a potential medical use, recent findings indicate that PXA might have an application as a research tool in the study of mitochondrial membrane dynamics, particularly non-canonical mitochondrial fission and remodelling of the mitochondrial matrix.[1]
Biological activity
[edit]Since PXA has antibiotic activity against organisms as diverse as bacteria, protozoans, fungi, plants and animal cells including human cancer cells, it has to affect a cellular feature that is evolutionarily highly conserved. A recent study has shown that PXA directly affects the mitochondria by disrupting both their biochemical functions and their membrane architecture.[1] The mitochondria are cellular organelles that are present in almost all eukaryotes. According to the theory of symbiogenesis, they are derived from bacteria and share many characteristics with them, including several properties of their membrane composition.[10][11]
One of the main functions of the mitochondria is to produce the cellular energy currency ATP through the process of oxidative phosphorylation (OxPhos). OxPhos depends on the mitochondrial membrane potential, which is generated by the electron transport chain (ETC) via the consumption of oxygen. PXA was shown to interfere with all of these functions of the mitochondria: not only does it decrease ATP synthesis and depolarise the mitochondria, but it also inhibits the ETC and cellular oxygen consumption. This sets it apart from uncoupling agents such as protonophores. While these also decrease ATP synthesis and depolarise the mitochondria, they increase respiration at the same time due to increased ETC activity in an attempt to restore the membrane potential.[1]
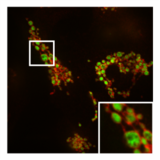
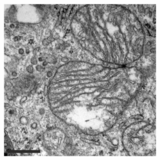
In addition to this inhibition of the function of mitochondria, PXA also disrupts their membrane architecture. In many cell types, the mitochondria normally form an intricate tubular network that undergoes a constant process of balanced mitochondrial fission and mitochondrial fusion. Treatment with PXA or many other mitochondrial stressors, such as protonophores, causes excessive fission that results in mitochondrial fragmentation. In the case of PXA, however, this fragmentation process was shown to be different from canonical fragmentation, caused by other agents such as protonophores, in several ways: first, it is considerably faster, resulting in complete fragmentation within a minute as opposed to about 30–60 minutes for canonical fragmentation; second, it is independent from the mitochondrial fission and fusion regulators DRP1 and OPA1; and third, while PXA causes fragmentation of both the outer mitochondrial membrane (OMM) and the mitochondrial matrix in wild type cells, it causes exclusive fragmentation of the matrix in cells that lack DRP1.[1] This last feature is especially unusual since no active mechanism for exclusive matrix fission is known in higher eukaryotes.[12] Examination of the mitochondrial ultrastructure revealed that PXA causes cristae disruption and complete distortion of the mitochondrial matrix. It is probably through this effect that PXA induces programmed cell death in the form of apoptosis.[1]
-
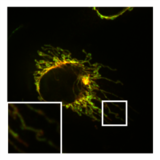
Normal, tubular morphology of mitochondria in a HeLa cell in which the fission mediator DRP1 has been knocked out. Overlay image of mitochondrial matrix (green) and OMM (red).
References
[edit]- ^ a b c d e f g h i j Böhler, Philip; Stuhldreier, Fabian; Anand, Ruchika; Kondadi, Arun Kumar; Schlütermann, David; Berleth, Niklas; Deitersen, Jana; Wallot-Hieke, Nora; Wu, Wenxian; Frank, Marian; Niemann, Hendrik; Wesbuer, Elisabeth; Barbian, Andreas; Luyten, Tomas; Parys, Jan B; Weidtkamp-Peters, Stefanie; Borchardt, Andrea; Reichert, Andreas S; Peña-Blanco, Aida; García-Sáez, Ana J; Itskanov, Samuel; Van Der Bliek, Alexander M; Proksch, Peter; Wesselborg, Sebastian; Stork, Björn (2018). "The mycotoxin phomoxanthone a disturbs the form and function of the inner mitochondrial membrane". Cell Death & Disease. 9 (3): 286. doi:10.1038/s41419-018-0312-8. PMC 5833434. PMID 29459714.
- ^ a b c d e Isaka, Masahiko; Jaturapat, Amonlaya; Rukseree, Kamolchanok; Danwisetkanjana, Kannawat; Tanticharoen, Morakot; Thebtaranonth, Yodhathai (2001). "Phomoxanthones A and B, Novel Xanthone Dimers from the Endophytic Fungus Phomopsis Species". Journal of Natural Products. 64 (8): 1015–8. doi:10.1021/np010006h. PMID 11520217. S2CID 32242541.
- ^ a b c d e Rönsberg, David; Debbab, Abdessamad; Mándi, Attila; Vasylyeva, Vera; Böhler, Philip; Stork, Björn; Engelke, Laura; Hamacher, Alexandra; Sawadogo, Richard; Diederich, Marc; Wray, Victor; Lin, Wenhan; Kassack, Matthias U; Janiak, Christoph; Scheu, Stefanie; Wesselborg, Sebastian; Kurtán, Tibor; Aly, Amal H; Proksch, Peter (2013). "Pro-Apoptotic and Immunostimulatory Tetrahydroxanthone Dimers from the Endophytic Fungus Phomopsis longicolla". Journal of Organic Chemistry. 78 (24): 12409–25. doi:10.1021/jo402066b. PMID 24295452.
- ^ Qin, Tian; Iwata, Takayuki; Ransom, Tanya T; Beutler, John A; Porco, John A (2015). "Syntheses of Dimeric Tetrahydroxanthones with Varied Linkages: Investigation of "Shapeshifting" Properties". Journal of the American Chemical Society. 137 (48): 15225–33. Bibcode:2015JAChS.13715225Q. doi:10.1021/jacs.5b09825. PMC 4863954. PMID 26544765.
- ^ Xing, X.K; Chen, J; Xu, M.J; Lin, W.H; Guo, S.X (2011). "Fungal endophytes associated with Sonneratia (Sonneratiaceae) mangrove plants on the south coast of China". Forest Pathology. 41 (4): 334. doi:10.1111/j.1439-0329.2010.00683.x.
- ^ Hobbs, Thomas W; Schmitthenner, A. F; Kuter, Geoffrey A (1985). "A New Phomopsis Species from Soybean". Mycologia. 77 (4): 535. doi:10.2307/3793352. JSTOR 3793352.
- ^ Li, Shuxian; Darwish, Omar; Alkharouf, Nadim W; Musungu, Bryan; Matthews, Benjamin F (2017). "Analysis of the genome sequence of Phomopsis longicolla: A fungal pathogen causing Phomopsis seed decay in soybean". BMC Genomics. 18 (1): 688. doi:10.1186/s12864-017-4075-x. PMC 5584002. PMID 28870170.
- ^ Elsässer, Brigitta; Krohn, Karsten; Flörke, Ulrich; Root, Natalia; Aust, Hans-Jürgen; Draeger, Siegfried; Schulz, Barbara; Antus, Sándor; Kurtán, Tibor (2005). "X-ray Structure Determination, Absolute Configuration and Biological Activity of Phomoxanthone A". European Journal of Organic Chemistry. 2005 (21): 4563. doi:10.1002/ejoc.200500265.
- ^ Frank, M; Niemann, H; Böhler, P; Stork, B; Wesselborg, S; Lin, W; Proksch, P (2015). "Phomoxanthone A--From Mangrove Forests to Anticancer Therapy". Current Medicinal Chemistry. 22 (30): 3523–32. doi:10.2174/0929867322666150716115300. PMID 26179997.
- ^ Martin, William F; Garg, Sriram; Zimorski, Verena (2015). "Endosymbiotic theories for eukaryote origin". Philosophical Transactions of the Royal Society B: Biological Sciences. 370 (1678): 20140330. doi:10.1098/rstb.2014.0330. PMC 4571569. PMID 26323761.
- ^ Mileykovskaya, E; Dowhan, W (2009). "Cardiolipin Membrane Domains in Prokaryotes and Eukaryotes". Biochimica et Biophysica Acta (BBA) - Biomembranes. 1788 (10): 2084–2091. doi:10.1016/j.bbamem.2009.04.003. PMC 2757463. PMID 19371718.
- ^ Van Der Bliek, A. M; Shen, Q; Kawajiri, S (2013). "Mechanisms of Mitochondrial Fission and Fusion". Cold Spring Harbor Perspectives in Biology. 5 (6): a011072. doi:10.1101/cshperspect.a011072. PMC 3660830. PMID 23732471.
External links
[edit] Media related to Phomoxanthone at Wikimedia Commons
Media related to Phomoxanthone at Wikimedia Commons