Multiple displacement amplification
Multiple displacement amplification (MDA) is a DNA amplification technique. This method can rapidly amplify minute amounts of DNA samples to a reasonable quantity for genomic analysis. The reaction starts by annealing random hexamer primers to the template: DNA synthesis is carried out by a high fidelity enzyme, preferentially Φ29 DNA polymerase. Compared with conventional PCR amplification techniques, MDA does not employ sequence-specific primers but amplifies all DNA, generates larger-sized products with a lower error frequency, and works at a constant temperature. MDA has been actively used in whole genome amplification (WGA) and is a promising method for application to single cell genome sequencing and sequencing-based genetic studies.
Background
[edit]Many biological and forensic cases involving genetic analysis require sequencing of DNA from minute amounts of sample, such as DNA from uncultured single cells or trace amounts of tissue collected from crime scenes. Conventional Polymerase Chain Reaction (PCR)-based DNA amplification methods require sequence-specific oligonucleotide primers and heat-stable (usually Taq) polymerase, and can be used to generate significant amounts of DNA from minute amounts of DNA. However, this is not sufficient for modern techniques which use sequencing-based DNA analysis. Therefore, a more efficient non-sequence-specific method to amplify minute amounts of DNA is necessary, especially in single-cell genomic studies.
Materials
[edit]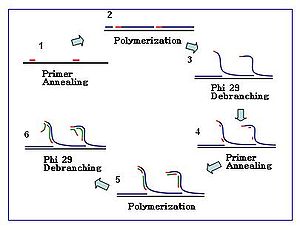
Phi 29 DNA polymerase
[edit]Bacteriophage Φ29 DNA polymerase is a high-processivity enzyme that can produce DNA amplicons greater than 70 kilobase pairs.[1] Its high fidelity and 3’–5' proofreading activity reduces the amplification error rate to 1 in 106−107 bases compared to conventional Taq polymerase with a reported error rate of 1 in 9,000.[2] The reaction can be carried out at a moderate isothermal condition of 30 °C and therefore does not require a thermocycler. It has been actively used in cell-free cloning, which is the enzymatic method of amplifying DNA in vitro without cell culturing and DNA extraction. The large fragment of Bst DNA polymerase is also used in MDA, but Ф29 is generally preferred due to its sufficient product yield and proofreading activity.[3]
Hexamer primers
[edit]Hexamer primers are sequences composed of six random nucleotides. For MDA applications, these primers are usually thiophosphate-modified at their 3’ end to convey resistance to the 3’–5’ exonuclease activity of Ф29 DNA polymerase. MDA reactions start with the annealing of such primers to the DNA template followed by polymerase-mediated chain elongation. Increasing numbers of primer annealing events happen along the amplification reaction.
Reaction
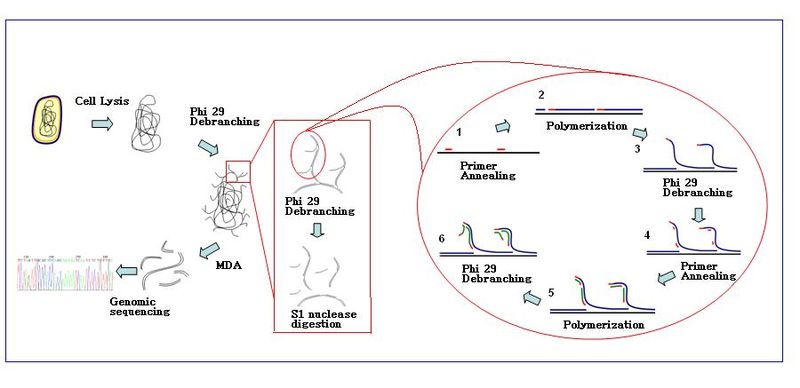
[edit]The amplification reaction initiates when multiple primer hexamers anneal to the template. When DNA synthesis proceeds to the next starting site, the polymerase displaces the newly produced DNA strand and continues its strand elongation. The strand displacement generates a newly synthesized single-stranded DNA template for more primers to anneal. Further primer annealing and strand displacement on the newly synthesized template results in a hyper-branched DNA network. The sequence debranching during amplification results in a high yield of the products. To separate the DNA branching network, S1 nucleases are used to cleave the fragments at displacement sites. The nicks on the resulting DNA fragments are repaired by DNA polymerase I.
Product quality
[edit]MDA can generate 1–2 μg of DNA from single cell with genome coverage of up to 99%.[4] Products also have lower error rate and larger sizes compared to PCR based Taq amplification.[4][5]
General work flow of MDA:[6]
- Sample preparation: Samples are collected and diluted in the appropriate reaction buffer (Ca2+ and Mg2+ free). Cells are lysed with alkaline buffer.
- Condition: The MDA reaction with Ф29 polymerase is carried out at 30 °C. The reaction usually takes about 2.5–3 hours.
- End of reaction: Inactivate enzymes at 65 °C before collection of the amplified DNA products
- DNA products can be purified with commercial purification kit.
Advantages
[edit]MDA generates sufficient yield of DNA products. It is a powerful tool of amplifying DNA molecules from samples, such as uncultured microorganism or single cells to the amount that would be sufficient for sequencing studies. The large size of MDA-amplified DNA products also provides desirable sample quality for identifying the size of polymorphic repeat alleles. Its high fidelity also makes it reliable to be used in the single-nucleotide polymorphism (SNP) allele detection. Due to its strand displacement during amplification, the amplified DNA has sufficient coverage of the source DNA molecules, which provides a high-quality product for genomic analysis. The products of displaced strands can be subsequently cloned into vectors to construct library for subsequent sequencing reactions.
Limitations
[edit]Allelic dropout (ADO)
[edit]ADO is defined as the random non-amplification of one of the alleles present in a heterozygous sample. Some studies have reported the ADO rate of the MDA products to be 0–60%.[7] This drawback decreases the accuracy of genotyping of single sample and misdiagnosis in other MDA involved applications. ADO appears to be independent of the fragment sizes and has been reported to have a similar rate in other single-cell techniques. Possible solutions are the use of different lysis conditions or to carry out multiple rounds of amplifications from the diluted MDA products since PCR mediated amplification from cultured cells has been reported to give lower ADO rates.
Preferential amplification
[edit]'Preferential amplification' is over-amplification of one of the alleles in comparison to the other. Most studies on MDA have reported this issue. The amplification bias is currently observed to be random. It might affect the analysis of small stretches of genomic DNA in identifying Short Tandem Repeats (STR) alleles.
Primer-primer interactions
[edit]Endogenous template-independent primer-primer interaction is due to the random design of hexamer primers. One possible solution is to design constrained-randomized hexanucleotide primers that do not cross-hybridize.
Applications
[edit]Single cell genome sequencing
[edit]Single cells of uncultured bacteria, archaea and protists, as well as individual viral particles and single fungal spores have been sequenced with the help of MDA.[8][9][10][11][12][13][14][15][16][17][18]
The ability to sequence individual cells is also useful in combating human disease. Genomes from single human embryonic cells have been successfully amplified for sequencing using MDA, allowing preimplantation genetic diagnosis (PGD): screening for genetic health issues in an early-stage embryo before implantation.[19] Diseases with heterogeneous properties, such as cancer, also benefit from MDA-based genome sequencing's ability to study mutations in individual cells.
The MDA products from a single cell have also been successfully used in array-comparative genomic hybridization experiments, which usually require a relatively large amount of amplified DNA.
Chromatin immunoprecipitation
[edit]Chromatin Immunoprecipitation results in production of complex mixtures of relatively short DNA fragments, which is challenging to amplify with MDA without causing a bias in the fragment representation. A method to circumvent this problem was proposed, which is based on conversion of these mixtures to circular concatemers using ligation, followed by Φ29 DNA polymerase-mediated MDA.[20]
Forensic analysis
[edit]The trace amount of samples collected from crime scenes can be amplified by MDA to the quantity that is enough for forensic DNA analysis, which is commonly used in identifying victims and suspects.
See also
[edit]References
[edit]- ^ Blanco L, Bernad A, Lázaro JM, Martín G, Garmendia C, Salas M (1989). "Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication". The Journal of Biological Chemistry. 264 (15): 8935–40. doi:10.1016/S0021-9258(18)81883-X. PMID 2498321.
- ^ Tindall KR and Kunkel TA (1988). "Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase". Biochemistry. 27 (16): 6008–13. doi:10.1021/bi00416a027. PMID 2847780.
- ^ Hutchison, C. A.; Smith, HO; Pfannkoch, C; Venter, JC (2005). "Cell-free cloning using φ29 DNA polymerase". Proceedings of the National Academy of Sciences. 102 (48): 17332–6. Bibcode:2005PNAS..10217332H. doi:10.1073/pnas.0508809102. PMC 1283157. PMID 16286637.
- ^ a b Paez JG, Lin M, Beroukhim R, Lee JC, Zhao X, Richter DJ, Gabriel S, Herman P, Sasaki H, Altshuler D, Li C, Meyerson M, Sellers WR (2004). "Genome coverage and sequence fidelity of phi29 polymerase-based multiple strand displacement whole genome amplification". Nucleic Acids Research. 32 (9): e71. doi:10.1093/nar/gnh069. PMC 419624. PMID 15150323.
- ^ Esteban JA, Salas M, Blanco L (1993). "Fidelity of phi 29 DNA polymerase. Comparison between protein-primed initiation and DNA polymerization". The Journal of Biological Chemistry. 268 (4): 2719–26. doi:10.1016/S0021-9258(18)53833-3. PMID 8428945.
- ^ Spits; Le Caignec, C; De Rycke, M; Van Haute, L; Van Steirteghem, A; Liebaers, I; Sermon, K (2006). "Whole-genome multiple displacement amplification from single cells". Nature Protocols. 1 (4): 1965–70. doi:10.1038/nprot.2006.326. PMID 17487184. S2CID 33346321.
- ^ Bradley, Ward, Yarborough (2012). "Allelic dropout rates in multiple displacement amplification". Applied Biomolecular Techniques. 84 (51): 341–362.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Zhang; Martiny, AC; Reppas, NB; Barry, KW; Malek, J; Chisholm, SW; Church, GM (2006). "Sequencing genomes from single cells by polymerase cloning". Nature Biotechnology. 24 (6): 680–6. doi:10.1038/nbt1214. PMID 16732271. S2CID 2994579.
- ^ Stepanauskas, Ramunas; Sieracki, Michael E. (2007-05-22). "Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time". Proceedings of the National Academy of Sciences. 104 (21): 9052–9057. Bibcode:2007PNAS..104.9052S. doi:10.1073/pnas.0700496104. ISSN 0027-8424. PMC 1885626. PMID 17502618.
- ^ Yoon, Hwan Su; Price, Dana C.; Stepanauskas, Ramunas; Rajah, Veeran D.; Sieracki, Michael E.; Wilson, William H.; Yang, Eun Chan; Duffy, Siobain; Bhattacharya, Debashish (2011-05-06). "Single-Cell Genomics Reveals Organismal Interactions in Uncultivated Marine Protists". Science. 332 (6030): 714–717. Bibcode:2011Sci...332..714Y. doi:10.1126/science.1203163. ISSN 0036-8075. PMID 21551060. S2CID 34343205.
- ^ Swan, Brandon K.; Martinez-Garcia, Manuel; Preston, Christina M.; Sczyrba, Alexander; Woyke, Tanja; Lamy, Dominique; Reinthaler, Thomas; Poulton, Nicole J.; Masland, E. Dashiell P. (2011-09-02). "Potential for Chemolithoautotrophy Among Ubiquitous Bacteria Lineages in the Dark Ocean". Science. 333 (6047): 1296–1300. Bibcode:2011Sci...333.1296S. doi:10.1126/science.1203690. ISSN 0036-8075. PMID 21885783. S2CID 206533092.
- ^ Woyke, Tanja; Xie, Gary; Copeland, Alex; González, José M.; Han, Cliff; Kiss, Hajnalka; Saw, Jimmy H.; Senin, Pavel; Yang, Chi (2009-04-23). "Assembling the Marine Metagenome, One Cell at a Time". PLOS ONE. 4 (4): e5299. Bibcode:2009PLoSO...4.5299W. doi:10.1371/journal.pone.0005299. ISSN 1932-6203. PMC 2668756. PMID 19390573.
- ^ Swan, Brandon K.; Tupper, Ben; Sczyrba, Alexander; Lauro, Federico M.; Martinez-Garcia, Manuel; González, José M.; Luo, Haiwei; Wright, Jody J.; Landry, Zachary C. (2013-07-09). "Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean". Proceedings of the National Academy of Sciences. 110 (28): 11463–11468. Bibcode:2013PNAS..11011463S. doi:10.1073/pnas.1304246110. ISSN 0027-8424. PMC 3710821. PMID 23801761.
- ^ Rinke, Christian; Schwientek, Patrick; Sczyrba, Alexander; Ivanova, Natalia N.; Anderson, Iain J.; Cheng, Jan-Fang; Darling, Aaron; Malfatti, Stephanie; Swan, Brandon K. (July 2013). "Insights into the phylogeny and coding potential of microbial dark matter". Nature. 499 (7459): 431–437. Bibcode:2013Natur.499..431R. doi:10.1038/nature12352. hdl:10453/27467. ISSN 0028-0836. PMID 23851394.
- ^ Kashtan, Nadav; Roggensack, Sara E.; Rodrigue, Sébastien; Thompson, Jessie W.; Biller, Steven J.; Coe, Allison; Ding, Huiming; Marttinen, Pekka; Malmstrom, Rex R. (2014-04-25). "Single-Cell Genomics Reveals Hundreds of Coexisting Subpopulations in Wild Prochlorococcus". Science. 344 (6182): 416–420. Bibcode:2014Sci...344..416K. doi:10.1126/science.1248575. hdl:1721.1/92763. ISSN 0036-8075. PMID 24763590. S2CID 13659345.
- ^ Wilson, William H; Gilg, Ilana C; Moniruzzaman, Mohammad; Field, Erin K; Koren, Sergey; LeCleir, Gary R; Martínez Martínez, Joaquín; Poulton, Nicole J; Swan, Brandon K (2017-05-12). "Genomic exploration of individual giant ocean viruses". The ISME Journal. 11 (8): 1736–1745. Bibcode:2017ISMEJ..11.1736W. doi:10.1038/ismej.2017.61. ISSN 1751-7362. PMC 5520044. PMID 28498373.
- ^ Stepanauskas, Ramunas; Fergusson, Elizabeth A.; Brown, Joseph; Poulton, Nicole J.; Tupper, Ben; Labonté, Jessica M.; Becraft, Eric D.; Brown, Julia M.; Pachiadaki, Maria G. (2017-07-20). "Improved genome recovery and integrated cell-size analyses of individual uncultured microbial cells and viral particles". Nature Communications. 8 (1): 84. Bibcode:2017NatCo...8...84S. doi:10.1038/s41467-017-00128-z. ISSN 2041-1723. PMC 5519541. PMID 28729688.
- ^ Pachiadaki, Maria G.; Sintes, Eva; Bergauer, Kristin; Brown, Julia M.; Record, Nicholas R.; Swan, Brandon K.; Mathyer, Mary Elizabeth; Hallam, Steven J.; Lopez-Garcia, Purificacion (2017-11-24). "Major role of nitrite-oxidizing bacteria in dark ocean carbon fixation". Science. 358 (6366): 1046–1051. Bibcode:2017Sci...358.1046P. doi:10.1126/science.aan8260. ISSN 0036-8075. PMID 29170234.
- ^ Coskun; Alsmadi, O (2007). "Whole genome amplification from a single cell: a new era for preimplantation genetic diagnosis". Prenatal Diagnosis. 27 (4): 297–302. doi:10.1002/pd.1667. PMID 17278176. S2CID 22175397.
- ^ Shoaib; Baconnais, S; Mechold, U; Le Cam, E; Lipinski, M; Ogryzko, V (2008). "Multiple displacement amplification for complex mixtures of DNA fragments". BMC Genomics. 9: 415. doi:10.1186/1471-2164-9-415. PMC 2553422. PMID 18793430.
