mtDNA control region
| Mitochondrial DNA control region secondary structure A | |
|---|---|
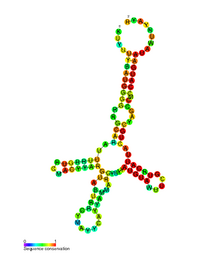 The consensus secondary structure for all haplotypes of the mtDNA control region. | |
| Identifiers | |
| Symbol | mtDNA ssA |
| Rfam | RF01853 |
| Other data | |
| RNA type | Antisense RNA |
| Domain(s) | Mammalia |
| PDB structures | PDBe |

The mtDNA control region is an area of the mitochondrial genome which is non-coding DNA. This region controls RNA and DNA synthesis.[1] It is the most polymorphic region of the human mtDNA genome,[2] with polymorphism concentrated in hypervariable regions. The average nucleotide diversity in these regions is 1.7%.[3] Despite this variability, an RNA transcript ("structure A") from this region has a conserved secondary structure (pictured) which has been found to be under selective pressure. There are 12 other secondary structures (structures B through M) in the human mtDNA control region with differing amounts of conservation.[4]
The mtDNA control region contains the origin of replication of one strand, and the origin of transcription for both strands.[5]
Distinction from D-loop
[edit]The control region and mtDNA D-loop are sometimes used synonymously in the literature;[3] specifically the control region includes the D-loop along with adjacent transcription promoter regions. For this reason, the control region is also known by the acronym DLP, standing for D-Loop and associated Promoters.[6]
D-loop means "displacement loop" and, in the context of mtDNA, specifically refers to a third strand that occurs as a copy of the heavy chain inside the NCR. Replication of mtDNA starts inside the D-loop.[7] The single displaced strand is also called 7S DNA. The primer used for 7S DNA synthesis is called 7S RNA.[8]
Endurance study
[edit]mtDNA control region haplotypes have been linked with endurance capacity in human subjects.[9] A 2002 study sequenced the control region of 55 subjects and compared their haplotype with the increase in VO2 max after an eight-week training program. They found that different haplotypes were significantly linked with the subjects' endurance. It was speculated that this was because the control region affects replication and transcription in the mitochondria.[4][9]
See also
[edit]References
[edit]- ^ Structure of the Mitochondrial Genome DNA Learning Center, Cold Spring Harbor Laboratory
- ^ Stoneking M, Hedgecock D, Higuchi RG, Vigilant L, Erlich HA (February 1991). "Population variation of human mtDNA control region sequences detected by enzymatic amplification and sequence-specific oligonucleotide probes". Am. J. Hum. Genet. 48 (2): 370–82. PMC 1683035. PMID 1990843.
- ^ a b Aquadro CF, Greenberg BD (February 1983). "Human Mitochondrial DNA Variation and Evolution: Analysis of Nucleotide Sequences from Seven Individuals". Genetics. 103 (2): 287–312. doi:10.1093/genetics/103.2.287. PMC 1219980. PMID 6299878. Retrieved 2010-07-29.
- ^ a b Pereira F, Soares P, Carneiro J, et al. (December 2008). "Evidence for variable selective pressures at a large secondary structure of the human mitochondrial DNA control region". Mol. Biol. Evol. 25 (12): 2759–70. doi:10.1093/molbev/msn225. PMID 18845547.
- ^ Anderson S, Bankier AT, Barrell BG, et al. (April 1981). "Sequence and organization of the human mitochondrial genome". Nature. 290 (5806): 457–65. Bibcode:1981Natur.290..457A. doi:10.1038/290457a0. PMID 7219534. S2CID 4355527.
- ^ Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G (October 1999). "Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication". Science. 286 (5440): 774–9. doi:10.1126/science.286.5440.774. PMID 10531063.
- ^ Fish, J.; Raule, N.; Attardi, G. (2004). "Discovery of a major D-loop replication origin reveals two modes of human mtDNA synthesis" (PDF). Science. 306 (5704): 2098–2101. Bibcode:2004Sci...306.2098F. doi:10.1126/science.1102077. PMID 15604407. S2CID 36033690.
- ^ Nicholls, Thomas J.; Minczuk, Michal (August 2014). "In D-loop: 40years of mitochondrial 7S DNA". Experimental Gerontology. 56: 175–181. doi:10.1016/j.exger.2014.03.027.
- ^ a b Murakami H, Ota A, Simojo H, Okada M, Ajisaka R, Kuno S (June 2002). "Polymorphisms in control region of mtDNA relates to individual differences in endurance capacity or trainability". Jpn. J. Physiol. 52 (3): 247–56. doi:10.2170/jjphysiol.52.247. PMID 12230801.
Further reading
[edit]- Stoneking M (October 2000). "Hypervariable Sites in the mtDNA Control Region Are Mutational Hotspots". Am. J. Hum. Genet. 67 (4): 1029–32. doi:10.1086/303092. PMC 1287875. PMID 10968778.
- Sachidanandam R, Weissman D, Schmidt SC, et al. (February 2001). "A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms". Nature. 409 (6822): 928–33. doi:10.1038/35057149. PMID 11237013.
External links
[edit]- EMPOP - the Mitochondrial DNA Control Region Database
- Page for Mitochondrial DNA control region secondary structure A at Rfam
