Non-mevalonate pathway
The non-mevalonate pathway—also appearing as the mevalonate-independent pathway and the 2-C-methyl-D-erythritol 4-phosphate/1-deoxy-D-xylulose 5-phosphate (MEP/DOXP) pathway—is an alternative metabolic pathway for the biosynthesis of the isoprenoid precursors isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP).[1][2][3] The currently preferred name for this pathway is the MEP pathway, since MEP is the first committed metabolite on the route to IPP.
Isoprenoid precursor biosynthesis
[edit]The mevalonate pathway (MVA pathway or HMG-CoA reductase pathway) and the MEP pathway are metabolic pathways for the biosynthesis of isoprenoid precursors: IPP and DMAPP. Whereas plants use both MVA and MEP pathway, most organisms only use one of the pathways for the biosynthesis of isoprenoid precursors. In plant cells IPP/DMAPP biosynthesis via the MEP pathway takes place in plastid organelles, while the biosynthesis via the MVA pathway takes place in the cytoplasm.[4] Most gram-negative bacteria, the photosynthetic cyanobacteria and green algae use only the MEP pathway.[5] Bacteria that use the MEP pathway include important pathogens such Mycobacterium tuberculosis.[6]
IPP and DMAPP serve as precursors for the biosynthesis of isoprenoid (terpenoid) molecules used in processes as diverse as protein prenylation, cell membrane maintenance, the synthesis of hormones, protein anchoring and N-glycosylation in all three domains of life.[citation needed] In photosynthetic organisms MEP-derived precursors are used for the biosynthesis of photosynthetic pigments, such as the carotenoids and the phytol chain of chlorophyll and light harvesting pigments.[5]
Bacteria such as Escherichia coli have been engineered for co-expressing biosynthesis genes of both the MEP and the MVA pathway.[7] Distribution of the metabolic fluxes between the MEP and the MVA pathway can be studied using 13C-glucose isotopomers.[8]
Reactions
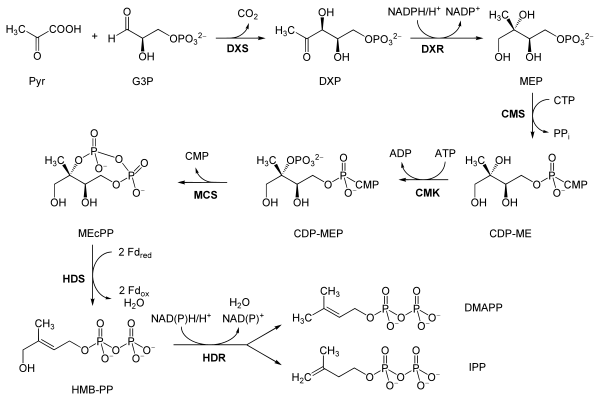
[edit]The reactions of the MEP pathway are as follows, taken primarily from Eisenreich and co-workers, except where the bold labels are additional local abbreviations to assist in connecting the table to the scheme above:[10][9]
| Reactants | Enzyme | Product | |
|---|---|---|---|
| Pyruvate (Pyr) and glyceraldehyde 3-phosphate (G3P) | DOXP synthase (Dxs; DXP) | 1-Deoxy-D-xylulose 5-phosphate (DOXP; DXP) |  |
| DOXP (DXP) | DXP reductoisomerase (Dxr, IspC; DXR) | 2-C-methylerythritol 4-phosphate (MEP) |  |
| MEP | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (YgbP, IspD; CMS) | 4-diphosphocytidyl-2-C-methylerythritol (CDP-ME) |  |
| CDP-ME | 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (YchB, IspE; CMK) | 4-diphosphocytidyl-2-C-methyl-D-erythritol 2-phosphate (CDP-MEP) | |
| CDP-MEP | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (YgbB, IspF; MCS) | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MEcPP) |  |
| MEcPP | HMB-PP synthase (GcpE, IspG; HDS) | (E)-4-Hydroxy-3-methyl-but-2-enyl pyrophosphate (HMB-PP) |  |
| HMB-PP | HMB-PP reductase (LytB, IspH; HDR) | Isopentenyl pyrophosphate (IPP) and Dimethylallyl pyrophosphate (DMAPP) |   |
Inhibition and other pathway research
[edit]Dxs, the first enzyme of the pathway, is feedback inhibited by the products IPP and DMAPP. Dxs is active as a homo-dimer and the precise mechanism of enzyme inhibition has been debated in the field. It has been proposed that IPP/DMAPP are competing the co-factor TPP.[11] A more recent study suggested that IPP/DMAPP trigger monomerisation and subsequent degradation of the enzyme, via interaction with a monomer interaction site that differs from the active site of the enzyme.[12]
DXP reductoisomerase (also known as: DXR, DOXP reductoisomerase, IspC, MEP synthase), is a key enzyme in the MEP pathway. It can be inhibited by the natural product fosmidomycin, which is under study as a starting point to develop a candidate antibacterial or antimalarial drug.[13][14][15]
The intermediate, HMB-PP, is a natural activator of human Vγ9/Vδ2 T cells, the major γδ T cell population in peripheral blood, and cells that "play a crucial role in the immune response to microbial pathogens".[16]
- IspH inhibitors: non-mevalonate Metabolic pathway that is essential for most bacteria but absent in humans, making it an ideal target for antibiotic development. This pathway, called methyl-D-erythritol phosphate (MEP) or non-mevalonate pathway, is responsible for biosynthesis of isoprenoids—molecules required for cell survival in most pathogenic bacteria and hence will be helpful in most usually antibacterial resistant bacteria.[17]
Metabolic engineering of the MEP/Non-mevalonate pathway
[edit]The MEP pathway has been extensively studied and engineered Escherichia coli, a commonly used microbial species for laboratory research and application.[18] IPP and DMAPP, the products of the MEP pathway can be used as substrates for the heterologous production of terpenoids with high value for application in the pharmaceutical and chemical industry. Upon expression of heterologous genes from different organisms, production of terpenoids like limonene, bisabolene and isoprene could be achieved in different microbial chassis.[19][20][21][22] Studies overexpressing different biosynthesis genes of the pathway revealed that expression of Dxs and Idi, catalyzing the first step and last step of the MEP pathway could significantly increase the yield of MEP derived terpenoids.[19][22] Dxs as the first enzyme of the pathway represents a bottleneck for the flux of carbon that enters the pathway. Idi which interconverts IPP to DMAPP and vice versa seems to be important for providing the respective substrate that is needed upon introduction of a heterologous carbon sink in engineered strains. A lot of metabolic engineering work on the MEP pathway has been done in cyanobacteria, photo-autotrophic microbes that can assimilate carbon dioxide from the atmosphere into various carbon containing metabolites, including terpenoids.[20][19][21] For biotechnology, cyanobacteria are, thus, an attractive platform for the sustainable production of high-value compounds.
References
[edit]- ^ Rohmer M; Rohmer, Michel (1999). "The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants". Nat Prod Rep. 16 (5): 565–574. doi:10.1039/a709175c. PMID 10584331.
- ^ W. Eisenreich; A. Bacher; D. Arigoni; F. Rohdich (2004). "Review Biosynthesis of isoprenoids via the non-mevalonate pathway". Cellular and Molecular Life Sciences. 61 (12): 1401–1426. doi:10.1007/s00018-004-3381-z. PMC 11138651. PMID 15197467. S2CID 24558920.
- ^ Hunter, WN (2007). "The Non-mevalonate Pathway of Isoprenoid Precursor Biosynthesis". Journal of Biological Chemistry. 282 (30): 21573–21577. doi:10.1074/jbc.R700005200. PMID 17442674.
- ^ Lichtenthaler H (1999). "The 1-Deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants". Annual Review of Plant Physiology and Plant Molecular Biology. 50: 47–65. doi:10.1146/annurev.arplant.50.1.47. PMID 15012203.
- ^ a b Vranová, Eva; Coman, Diana; Gruissem, Wilhelm (2013-04-29). "Network Analysis of the MVA and MEP Pathways for Isoprenoid Synthesis". Annual Review of Plant Biology. 64 (1): 665–700. doi:10.1146/annurev-arplant-050312-120116. ISSN 1543-5008. PMID 23451776.
- ^ Wiemer, AJ; Hsiao, CH; Wiemer, DF (2010). "Isoprenoid Metabolism as a Therapeutic Target in Gram-Negative Pathogens". Current Topics in Medicinal Chemistry. 10 (18): 1858–1871. doi:10.2174/156802610793176602. PMID 20615187.
- ^ Martin MJ, Pitera DJ, Withers ST, Newman JD, Keasling JD (2003). "Engineering a mevalonate pathway in Escherichia coli for production of terpenoids". Nature Biotechnology. 21 (7): 796–802. doi:10.1038/nbt833. PMID 12778056. S2CID 17214504.
- ^ Orsi E, Beekwilder J, Peek S, Eggink G, Kengen SW, Weusthuis RA (2020). "Metabolic flux ratio analysis by parallel 13C labeling of isoprenoid biosynthesis in Rhodobacter sphaeroides". Metabolic Engineering. 57: 228–238. doi:10.1016/j.ymben.2019.12.004. PMID 31843486.
- ^ a b Qidwai T, Jamal F, Khan MY, Sharma B (2014). "Exploring Drug Targets in Isoprenoid Biosynthetic Pathway for Plasmodium falciparum". Biochemistry Research International. 2014: 657189. doi:10.1155/2014/657189. PMC 4017727. PMID 24864210. (Retracted, see doi:10.1155/2022/8426183, PMID 35340427)
- ^ a b Eisenreich W, Bacher A, Arigoni D, Rohdich F (2004). "Biosynthesis of Isoprenoids Via the Non-mevalonate Pathway". Cell. Mol. Life Sci. 61 (12): 1401–26. doi:10.1007/s00018-004-3381-z. PMC 11138651. PMID 15197467. S2CID 24558920.
- ^ Banerjee, Aparajita; Wu, Yan; Banerjee, Rahul; Li, Yue; Yan, Honggao; Sharkey, Thomas D. (June 2013). "Feedback Inhibition of Deoxy-d-xylulose-5-phosphate Synthase Regulates the Methylerythritol 4-Phosphate Pathway". Journal of Biological Chemistry. 288 (23): 16926–16936. doi:10.1074/jbc.m113.464636. ISSN 0021-9258. PMC 3675625. PMID 23612965.
- ^ Di, Xueni; Ortega-Alarcon, David; Kakumanu, Ramu; Iglesias-Fernandez, Javier; Diaz, Lucia; Baidoo, Edward E. K.; Velazquez-Campoy, Adrian; Rodríguez-Concepción, Manuel; Perez-Gil, Jordi (2023-05-08). "MEP pathway products allosterically promote monomerization of deoxy-D-xylulose-5-phosphate synthase to feedback-regulate their supply". Plant Communications. 4 (3): 100512. doi:10.1016/j.xplc.2022.100512. hdl:10261/335525. ISSN 2590-3462. PMID 36575800.
- ^ Hale I, O'Neill PM, Berry NG, Odom A, Sharma R (2012). "The MEP pathway and the Development of Inhibitors as Potential Anti-Infective Agents". Med. Chem. Commun. 3 (4): 418–433. doi:10.1039/C2MD00298A.
- ^ Jomaa H, Wiesner J, Sanderbrand S, et al. (1999). "Inhibitors of the Nonmevalonate Pathway of Isoprenoid Biosynthesis as Antimalarial Drugs". Science. 285 (5433): 1573–6. doi:10.1126/science.285.5433.1573. PMID 10477522.
- ^ C. Zinglé; L. Kuntz; D. Tritsch; C. Grosdemange-Billiard; M. Rohmer (2010). "Isoprenoid Biosynthesis via the Methylerythritol Phosphate Pathway: Structural Variations around Phosphonate Anchor and Spacer of Fosmidomycin, a Potent Inhibitor of Deoxyxylulose Phosphate Reductoisomerase". J. Org. Chem. 75 (10): 3203–3207. doi:10.1021/jo9024732. PMID 20429517.
- ^ Eberl M, Hintz M, Reichenberg A, Kollas AK, Wiesner J, Jomaa H (2003). "Microbial Isoprenoid Biosynthesis and Human γδ T cell Activation". FEBS Lett. 544 (1–3): 4–10. doi:10.1016/S0014-5793(03)00483-6. PMID 12782281. S2CID 9930822.
- ^ "Research team reports new class of antibiotics active against a wide range of bacteria". MDLinx. 23 December 2020. Retrieved 23 January 2023.[dead link]
- ^ Rodrı́guez-Concepción, Manuel; Boronat, Albert (2002-11-01). "Elucidation of the Methylerythritol Phosphate Pathway for Isoprenoid Biosynthesis in Bacteria and Plastids. A Metabolic Milestone Achieved through Genomics". Plant Physiology. 130 (3): 1079–1089. doi:10.1104/pp.007138. PMC 1540259. PMID 12427975.
- ^ a b c Englund, Elias; Shabestary, Kiyan; Hudson, Elton P.; Lindberg, Pia (2018-09-01). "Systematic overexpression study to find target enzymes enhancing production of terpenes in Synechocystis PCC 6803, using isoprene as a model compound". Metabolic Engineering. 49: 164–177. doi:10.1016/j.ymben.2018.07.004. ISSN 1096-7176. PMID 30025762.
- ^ a b Gao, Xiang; Gao, Fang; Liu, Deng; Zhang, Hao; Nie, Xiaoqun; Yang, Chen (2016-04-13). "Engineering the methylerythritol phosphate pathway in cyanobacteria for photosynthetic isoprene production from CO2". Energy & Environmental Science. 9 (4): 1400–1411. doi:10.1039/C5EE03102H. ISSN 1754-5706.
- ^ a b Rodrigues, João S.; Lindberg, Pia (2021-06-01). "Metabolic engineering of Synechocystis sp. PCC 6803 for improved bisabolene production". Metabolic Engineering Communications. 12: e00159. doi:10.1016/j.mec.2020.e00159. ISSN 2214-0301. PMC 7809396. PMID 33489752.
- ^ a b Du, Fu-Liang; Yu, Hui-Lei; Xu, Jian-He; Li, Chun-Xiu (2014-08-31). "Enhanced limonene production by optimizing the expression of limonene biosynthesis and MEP pathway genes in E. coli". Bioresources and Bioprocessing. 1 (1): 10. doi:10.1186/s40643-014-0010-z. ISSN 2197-4365.