Gut microbiota

Gut microbiota, gut microbiome, or gut flora are the microorganisms, including bacteria, archaea, fungi, and viruses, that live in the digestive tracts of animals.[1][2] The gastrointestinal metagenome is the aggregate of all the genomes of the gut microbiota.[3][4] The gut is the main location of the human microbiome.[5] The gut microbiota has broad impacts, including effects on colonization, resistance to pathogens, maintaining the intestinal epithelium, metabolizing dietary and pharmaceutical compounds, controlling immune function, and even behavior through the gut–brain axis.[4]
The microbial composition of the gut microbiota varies across regions of the digestive tract. The colon contains the highest microbial density of any human-associated microbial community studied so far, representing between 300 and 1000 different species.[6] Bacteria are the largest and to date, best studied component and 99% of gut bacteria come from about 30 or 40 species.[7] Up to 60% of the dry mass of feces is bacteria.[8] Over 99% of the bacteria in the gut are anaerobes, but in the cecum, aerobic bacteria reach high densities.[5] It is estimated that the human gut microbiota have around a hundred times as many genes as there are in the human genome.
Overview
[edit]
In humans, the gut microbiota has the highest numbers and species of bacteria compared to other areas of the body.[9] The approximate number of bacteria composing the gut microbiota is about 1013–1014 (10,000 to 100,000 billion).[10] In humans, the gut flora is established at birth and gradually transitions towards a state resembling that of adults by the age of two,[11] coinciding with the development and maturation of the intestinal epithelium and intestinal mucosal barrier. This barrier is essential for supporting a symbiotic relationship with the gut flora while providing protection against pathogenic organisms.[12][13]
The relationship between some gut microbiota and humans is not merely commensal (a non-harmful coexistence), but rather a mutualistic relationship.[5]: 700 Some human gut microorganisms benefit the host by fermenting dietary fiber into short-chain fatty acids (SCFAs), such as acetic acid and butyric acid, which are then absorbed by the host.[9][14] Intestinal bacteria also play a role in synthesizing certain B vitamins and vitamin K as well as metabolizing bile acids, sterols, and xenobiotics.[5][14] The systemic importance of the SCFAs and other compounds they produce are like hormones and the gut flora itself appears to function like an endocrine organ.[14] Dysregulation of the gut flora has been correlated with a host of inflammatory and autoimmune conditions.[9][15]
The composition of human gut microbiota changes over time, when the diet changes, and as overall health changes.[9][15] A systematic review from 2016 examined the preclinical and small human trials that have been conducted with certain commercially available strains of probiotic bacteria and identified those that had the most potential to be useful for certain central nervous system disorders.[16] It should also be highlighted that the Mediterranean diet, rich in vegetables and fibers, stimulates the activity and growth of beneficial bacteria for the brain.[17]
Classifications
[edit]The microbial composition of the gut microbiota varies across the digestive tract. In the stomach and small intestine, relatively few species of bacteria are generally present.[6][18] The colon, in contrast, contains the highest microbial density of any human-associated microbial community studied so far[19] with between 1010 and 1011 (10 to 100 billion) cells per gram of intestinal content.[20] These bacteria represent between 300 and 1000 different species.[6][18] However, 99% of the bacteria come from about 30 or 40 species.[7] As a consequence of their abundance in the intestine, bacteria also make up to 60% of the dry mass of feces.[8] Fungi, protists, archaea, and viruses are also present in the gut flora, but less is known about their activities.[21]
Over 99% of the bacteria in the gut are anaerobes, but in the cecum, aerobic bacteria reach high densities.[5] It is estimated that these gut flora have around a hundred times as many genes in total as there are in the human genome.[22]
Many species in the gut have not been studied outside of their hosts because they cannot be cultured.[18][7][23] While there are a small number of core microbial species shared by most individuals, populations of microbes can vary widely.[24] Within an individual, their microbial populations stay fairly constant over time, with some alterations occurring due to changes in lifestyle, diet and age.[6][25] The Human Microbiome Project has set out to better describe the microbiota of the human gut and other body locations.[citation needed]
The four dominant bacterial phyla in the human gut are Bacillota (Firmicutes), Bacteroidota, Actinomycetota, and Pseudomonadota.[26] Most bacteria belong to the genera Bacteroides, Clostridium, Faecalibacterium,[6][7] Eubacterium, Ruminococcus, Peptococcus, Peptostreptococcus, and Bifidobacterium.[6][7] Other genera, such as Escherichia and Lactobacillus, are present to a lesser extent.[6] Species from the genus Bacteroides alone constitute about 30% of all bacteria in the gut, suggesting that this genus is especially important in the functioning of the host.[18]
Fungal genera that have been detected in the gut include Candida, Saccharomyces, Aspergillus, Penicillium, Rhodotorula, Trametes, Pleospora, Sclerotinia, Bullera, and Galactomyces, among others.[27][28] Rhodotorula is most frequently found in individuals with inflammatory bowel disease while Candida is most frequently found in individuals with hepatitis B cirrhosis and chronic hepatitis B.[27]
Due to the prevalence of fungi in the natural environment, determining which genera and species are permanent members of the gut mycobiome is difficult.[29][30] Research is underway as to whether Penicillium is a permanent or transient member of the gut flora, obtained from dietary sources such as cheese, though several species in the genus are known to survive at temperatures around 37°C, around the same as the core body temperature.[30] Saccharomyces cerevisiae, brewer's yeast, is known to reach the intestines after being ingested and can be responsible for the condition auto-brewery syndrome in cases where it is overabundant,[30][31][32] while Candida albicans is likely a permanent member, and is believed to be acquired at birth through vertical transmission.[33][medical citation needed]
Archaea constitute another large class of gut flora which are important in the metabolism of the bacterial products of fermentation.
Industrialization is associated with changes in the microbiota and the reduction of diversity could drive certain species to extinction; in 2018, researchers proposed a biobank repository of human microbiota.[34]
Enterotype
[edit]An enterotype is a classification of living organisms based on its bacteriological ecosystem in the human gut microbiome not dictated by age, gender, body weight, or national divisions.[35] There are indications that long-term diet influences enterotype.[36] Three human enterotypes have been proposed,[35][37] but their value has been questioned.[38]
Composition
[edit]Bacteriome
[edit]Stomach
[edit]Due to the high acidity of the stomach, most microorganisms cannot survive there. The main bacteria of the gastric microbiota belong to five major phyla: Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteriota, and Proteobacteria. The dominant genera are Prevotella, Streptococcus, Veillonella, Rothia , and Haemophilus.[39] The interaction between the pre-existing gastric microbiota with the introduction of H. pylori may influence disease progression.[39] When there is a presence of H. pylori it becomes the dominant of the microbiota.[40]
Intestines
[edit]| Bacteria commonly found in the human colon[41] | |
| Bacterium | Incidence (%) |
|---|---|
| Bacteroides fragilis | 100 |
| Bacteroides melaninogenicus | 100 |
| Bacteroides oralis | 100 |
| Enterococcus faecalis | 100 |
| Escherichia coli | 100 |
| Enterobacter sp. | 40–80 |
| Klebsiella sp. | 40–80 |
| Bifidobacterium bifidum | 30–70 |
| Staphylococcus aureus | 30–50 |
| Lactobacillus | 20–60 |
| Clostridium perfringens | 25–35 |
| Proteus mirabilis | 5–55 |
| Clostridium tetani | 1–35 |
| Clostridium septicum | 5–25 |
| Pseudomonas aeruginosa | 3–11 |
| Salmonella enterica | 3–7 |
| Faecalibacterium prausnitzii | ?common |
| Peptostreptococcus sp. | ?common |
| Peptococcus sp. | ?common |
The small intestine contains a trace amount of microorganisms due to the proximity and influence of the stomach. Gram-positive cocci and rod-shaped bacteria are the predominant microorganisms found in the small intestine.[5] However, in the distal portion of the small intestine alkaline conditions support gram-negative bacteria of the Enterobacteriaceae.[5] The bacterial flora of the small intestine aid in a wide range of intestinal functions. The bacterial flora provide regulatory signals that enable the development and utility of the gut. Overgrowth of bacteria in the small intestine can lead to intestinal failure.[42] In addition the large intestine contains the largest bacterial ecosystem in the human body.[5] About 99% of the large intestine and feces flora are made up of obligate anaerobes such as Bacteroides and Bifidobacterium.[43] Factors that disrupt the microorganism population of the large intestine include antibiotics, stress, and parasites.[5]
Bacteria make up most of the flora in the colon[44] and accounts for 60% of fecal nitrogen.[6] This fact makes feces an ideal source of gut flora for any tests and experiments by extracting the nucleic acid from fecal specimens, and bacterial 16S rRNA gene sequences are generated with bacterial primers. This form of testing is also often preferable to more invasive techniques, such as biopsies.
Five phyla dominate the intestinal microbiota: Bacteroidota, Bacillota (Firmicutes), Actinomycetota, Pseudomonadota, and Verrucomicrobiota – with Bacteroidota and Bacillota constituting 90% of the composition.[45] Somewhere between 300[6] and 1000 different species live in the gut,[18] with most estimates at about 500.[46][47] However, it is probable that 99% of the bacteria come from about 30 or 40 species, with Faecalibacterium prausnitzii (phylum firmicutes) being the most common species in healthy adults.[7][48]
Research suggests that the relationship between gut flora and humans is not merely commensal (a non-harmful coexistence), but rather is a mutualistic, symbiotic relationship.[18] Though people can survive with no gut flora,[46] the microorganisms perform a host of useful functions, such as fermenting unused energy substrates, training the immune system via end products of metabolism like propionate and acetate, preventing growth of harmful species, regulating the development of the gut, producing vitamins for the host (such as biotin and vitamin K), and producing hormones to direct the host to store fats.[5] Extensive modification and imbalances of the gut microbiota and its microbiome or gene collection are associated with obesity.[49] However, in certain conditions, some species are thought to be capable of causing disease by causing infection or increasing cancer risk for the host.[6][44]
Mycobiome
[edit]Fungi and protists also make up a part of the gut flora, but less is known about their activities.[50]
Virome
[edit]The human virome is mostly bacteriophages.[51]
Variation
[edit]Age
[edit]There are common patterns of microbiome composition evolution during life.[52] In general, the diversity of microbiota composition of fecal samples is significantly higher in adults than in children, although interpersonal differences are higher in children than in adults.[53] Much of the maturation of microbiota into an adult-like configuration happens during the first three years of life.[53]
As the microbiome composition changes, so does the composition of bacterial proteins produced in the gut. In adult microbiomes, a high prevalence of enzymes involved in fermentation, methanogenesis and the metabolism of arginine, glutamate, aspartate and lysine have been found. In contrast, in infant microbiomes the dominant enzymes are involved in cysteine metabolism and fermentation pathways.[53]
Geography
[edit]Gut microbiome composition depends on the geographic origin of populations. Variations in a trade-off of Prevotella, the representation of the urease gene, and the representation of genes encoding glutamate synthase/degradation or other enzymes involved in amino acids degradation or vitamin biosynthesis show significant differences between populations from the US, Malawi, or Amerindian origin.[53]
The US population has a high representation of enzymes encoding the degradation of glutamine and enzymes involved in vitamin and lipoic acid biosynthesis; whereas Malawi and Amerindian populations have a high representation of enzymes encoding glutamate synthase and they also have an overrepresentation of α-amylase in their microbiomes. As the US population has a diet richer in fats than Amerindian or Malawian populations which have a corn-rich diet, the diet is probably the main determinant of the gut bacterial composition.[53]
Further studies have indicated a large difference in the composition of microbiota between European and rural African children. The fecal bacteria of children from Florence were compared to that of children from the small rural village of Boulpon in Burkina Faso. The diet of a typical child living in this village is largely lacking in fats and animal proteins and rich in polysaccharides and plant proteins. The fecal bacteria of European children were dominated by Firmicutes and showed a marked reduction in biodiversity, while the fecal bacteria of the Boulpon children was dominated by Bacteroidetes. The increased biodiversity and different composition of the gut microbiome in African populations may aid in the digestion of normally indigestible plant polysaccharides and also may result in a reduced incidence of non-infectious colonic diseases.[54]
On a smaller scale, it has been shown that sharing numerous common environmental exposures in a family is a strong determinant of individual microbiome composition. This effect has no genetic influence and it is consistently observed in culturally different populations.[53]
Malnourishment
[edit]Malnourished children have less mature and less diverse gut microbiota than healthy children, and changes in the microbiome associated with nutrient scarcity can in turn be a pathophysiological cause of malnutrition.[55][56] Malnourished children also typically have more potentially pathogenic gut flora, and more yeast in their mouths and throats.[57] Altering diet may lead to changes in gut microbiota composition and diversity.[58]
Race and ethnicity
[edit]Researchers with the American Gut Project and Human Microbiome Project found that twelve microbe families varied in abundance based on the race or ethnicity of the individual. The strength of these associations is limited by the small sample size: the American Gut Project collected data from 1,375 individuals, 90% of whom were white.[59] The Healthy Life in an Urban Setting (HELIUS) study in Amsterdam found that those of Dutch ancestry had the highest level of gut microbiota diversity, while those of South Asian and Surinamese descent had the lowest diversity. The study results suggested that individuals of the same race or ethnicity have more similar microbiomes than individuals of different racial backgrounds.[59]
Socioeconomic status
[edit]As of 2020, at least two studies have demonstrated a link between an individual's socioeconomic status (SES) and their gut microbiota. A study in Chicago found that individuals in higher SES neighborhoods had greater microbiota diversity. People from higher SES neighborhoods also had more abundant Bacteroides bacteria. Similarly, a study of twins in the United Kingdom found that higher SES was also linked with a greater gut diversity.[59]
Antibiotic Usage
[edit]As of 2023, a study suggests that antibiotics, especially those used in the treatment of broad-spectrum bacterial infections, have negative effects on the gut microbiota.[60] The study also states that there are many experts on intestinal health concerned that antibody usage has reduced the diversity of the gut microbiota, many of the strains are lost, and if there is a re-emergence of the bacteria, is gradual and long-term. [61]
Acquisition in human infants
[edit]The establishment of a gut flora is crucial to the health of an adult, as well as the functioning of the gastrointestinal tract.[62] In humans, a gut flora similar to an adult's is formed within one to two years of birth as microbiota are acquired through parent-to-child transmission and transfer from food, water, and other environmental sources.[63][12]

The traditional view of the gastrointestinal tract of a normal fetus is that it is sterile, although this view has been challenged in the past few years.[timeframe?][64] Multiple lines of evidence have begun to emerge that suggest there may be bacteria in the intrauterine environment. In humans, research has shown that microbial colonization may occur in the fetus[65] with one study showing Lactobacillus and Bifidobacterium species were present in placental biopsies.[66] Several rodent studies have demonstrated the presence of bacteria in the amniotic fluid and placenta, as well as in the meconium of babies born by sterile cesarean section.[67][68] In another study, researchers administered a culture of bacteria orally to pregnant mice, and detected the bacteria in the offspring, likely resulting from transmission between the digestive tract and amniotic fluid via the blood stream.[69] However, researchers caution that the source of these intrauterine bacteria, whether they are alive, and their role, is not yet understood.[70][66]
During birth and rapidly thereafter, bacteria from the mother and the surrounding environment colonize the infant's gut.[12] The exact sources of bacteria are not fully understood, but may include the birth canal, other people (parents, siblings, hospital workers), breastmilk, food, and the general environment with which the infant interacts.[71] Research has shown that the microbiome of babies born vaginally differs significantly from that of babies delivered by caesarean section and that vaginally born babies got most of their gut bacteria from their mother, while the microbiota of babies born by caesarean section had more bacteria associated with hospital environments.[72]
During the first year of life, the composition of the gut flora is generally simple and changes a great deal with time and is not the same across individuals.[12] The initial bacterial population are generally facultative anaerobic organisms; investigators believe that these initial colonizers decrease the oxygen concentration in the gut, which in turn allows obligately anaerobic bacteria like Bacteroidota, Actinomycetota, and Bacillota to become established and thrive.[12] Breast-fed babies become dominated by bifidobacteria, possibly due to the contents of bifidobacterial growth factors in breast milk, and by the fact that breast milk carries prebiotic components, allowing for healthy bacterial growth.[66][73] Breast milk also contains higher levels of Immunoglobulin A (IgA) to help with the tolerance and regulation of the baby's immune system.[74] In contrast, the microbiota of formula-fed infants is more diverse, with high numbers of Enterobacteriaceae, enterococci, bifidobacteria, Bacteroides, and clostridia.[75]
Caesarean section, antibiotics, and formula feeding may alter the gut microbiome composition.[66] Children treated with antibiotics have less stable, and less diverse floral communities.[76] Caesarean sections have been shown to be disruptive to mother-offspring transmission of bacteria, which impacts the overall health of the offspring by raising risks of disease such as celiac disease, asthma, and type 1 diabetes.[66] This further evidences the importance of a healthy gut microbiome. Various methods of microbiome restoration are being explored, typically involving exposing the infant to maternal vaginal contents, and oral probiotics.[66]
Functions
[edit]When the study of gut flora began in 1995,[77] it was thought to have three key roles: direct defense against pathogens, fortification of host defense by its role in developing and maintaining the intestinal epithelium and inducing antibody production there, and metabolizing otherwise indigestible compounds in food. Subsequent work discovered its role in training the developing immune system, and yet further work focused on its role in the gut–brain axis.[78]
Direct inhibition of pathogens
[edit]The gut flora community plays a direct role in defending against pathogens by fully colonising the space, making use of all available nutrients, and by secreting compounds known as cytokines that kill or inhibit unwelcome organisms that would compete for nutrients with it.[79] Different strains of gut bacteria cause the production of different cytokines. Cytokines are chemical compounds produced by our immune system for initiating the inflammatory response against infections. Disruption of the gut flora allows competing organisms like Clostridioides difficile to become established that otherwise are kept in abeyance.[79]
Development of enteric protection and immune system
[edit]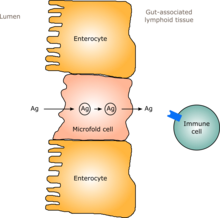
In humans, a gut flora similar to an adult's is formed within one to two years of birth.[12] As the gut flora gets established, the lining of the intestines – the intestinal epithelium and the intestinal mucosal barrier that it secretes – develop as well, in a way that is tolerant to, and even supportive of, commensalistic microorganisms to a certain extent and also provides a barrier to pathogenic ones.[12] Specifically, goblet cells that produce the mucosa proliferate, and the mucosa layer thickens, providing an outside mucosal layer in which "friendly" microorganisms can anchor and feed, and an inner layer that even these organisms cannot penetrate.[12][13] Additionally, the development of gut-associated lymphoid tissue (GALT), which forms part of the intestinal epithelium and which detects and reacts to pathogens, appears and develops during the time that the gut flora develops and established.[12] The GALT that develops is tolerant to gut flora species, but not to other microorganisms.[12] GALT also normally becomes tolerant to food to which the infant is exposed, as well as digestive products of food, and gut flora's metabolites (molecules formed from metabolism) produced from food.[12]
The human immune system creates cytokines that can drive the immune system to produce inflammation in order to protect itself, and that can tamp down the immune response to maintain homeostasis and allow healing after insult or injury.[12] Different bacterial species that appear in gut flora have been shown to be able to drive the immune system to create cytokines selectively; for example Bacteroides fragilis and some Clostridia species appear to drive an anti-inflammatory response, while some segmented filamentous bacteria drive the production of inflammatory cytokines.[12][80] Gut flora can also regulate the production of antibodies by the immune system.[12][81] One function of this regulation is to cause B cells to class switch to IgA. In most cases B cells need activation from T helper cells to induce class switching; however, in another pathway, gut flora cause NF-kB signaling by intestinal epithelial cells which results in further signaling molecules being secreted.[82] These signaling molecules interact with B cells to induce class switching to IgA.[82] IgA is an important type of antibody that is used in mucosal environments like the gut. It has been shown that IgA can help diversify the gut community and helps in getting rid of bacteria that cause inflammatory responses.[83] Ultimately, IgA maintains a healthy environment between the host and gut bacteria.[83] These cytokines and antibodies can have effects outside the gut, in the lungs and other tissues.[12]
The immune system can also be altered due to the gut bacteria's ability to produce metabolites that can affect cells in the immune system. For example short-chain fatty acids (SCFA) can be produced by some gut bacteria through fermentation.[84] SCFAs stimulate a rapid increase in the production of innate immune cells like neutrophils, basophils and eosinophils.[84] These cells are part of the innate immune system that try to limit the spread of infection.
Metabolism
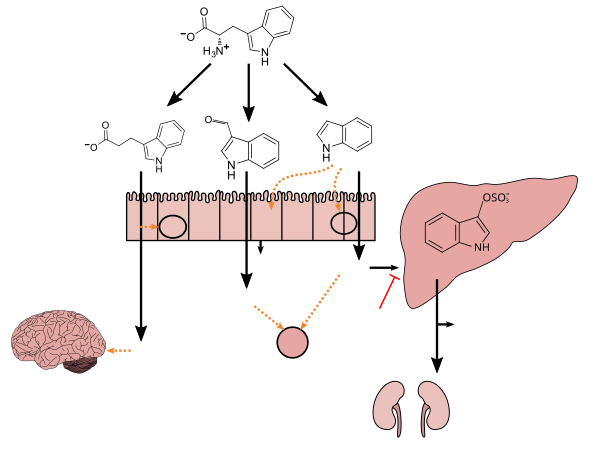
[edit]Tryptophan metabolism by human gut microbiota ()
|
Without gut flora, the human body would be unable to utilize some of the undigested carbohydrates it consumes, because some types of gut flora have enzymes that human cells lack for breaking down certain polysaccharides.[14] Rodents raised in a sterile environment and lacking in gut flora need to eat 30% more calories just to remain the same weight as their normal counterparts.[14] Carbohydrates that humans cannot digest without bacterial help include certain starches, fiber, oligosaccharides, and sugars that the body failed to digest and absorb like lactose in the case of lactose intolerance and sugar alcohols, mucus produced by the gut, and proteins.[9][14]
Bacteria turn carbohydrates they ferment into short-chain fatty acids by a form of fermentation called saccharolytic fermentation.[47] Products include acetic acid, propionic acid and butyric acid.[7][47] These materials can be used by host cells, providing a major source of energy and nutrients.[47] Gases (which are involved in signaling[89] and may cause flatulence) and organic acids, such as lactic acid, are also produced by fermentation.[7] Acetic acid is used by muscle, propionic acid facilitates liver production of ATP, and butyric acid provides energy to gut cells.[47]
Gut flora also synthesize vitamins like biotin and folate, and facilitate absorption of dietary minerals, including magnesium, calcium, and iron.[6][25] Methanobrevibacter smithii is unique because it is not a species of bacteria, but rather a member of domain Archaea, and is the most abundant methane-producing archaeal species in the human gastrointestinal microbiota.[90]
Gut microbiota also serve as a source of vitamins K and B12, which are not produced by the body or produced in little amount.[91][92]
Cellulose degradation
[edit]Bacteria that degrade cellulose (such as Ruminococcus) are prevalent among great apes, ancient human societies, hunter-gatherer communities, and even modern rural populations. However, they are rare in industrialized societies. Human-associated strains have acquired genes that can degrade specific plant fibers such as maize, rice, and wheat. Bacterial strains found in primates can also degrade chitin, a polymer abundant in insects, which are part of the diet of many nonhuman primates. The decline of these bacteria in the human gut were likely influenced by the shift toward western lifestyles.[93]
Pharmacomicrobiomics
[edit]The human metagenome (i.e., the genetic composition of an individual and all microorganisms that reside on or within the individual's body) varies considerably between individuals.[94][95] Since the total number of microbial cells in the human body (over 100 trillion) greatly outnumbers Homo sapiens cells (tens of trillions),[note 1][94][96] there is considerable potential for interactions between drugs and an individual's microbiome, including: drugs altering the composition of the human microbiome, drug metabolism by microbial enzymes modifying the drug's pharmacokinetic profile, and microbial drug metabolism affecting a drug's clinical efficacy and toxicity profile.[94][95][97]
Apart from carbohydrates, gut microbiota can also metabolize other xenobiotics such as drugs, phytochemicals, and food toxicants. More than 30 drugs have been shown to be metabolized by gut microbiota.[98] The microbial metabolism of drugs can sometimes inactivate the drug.[99]
Contribution to drug metabolism
[edit]The gut microbiota is an enriched community that contains diverse genes with huge biochemical capabilities to modify drugs, especially those taken by mouth.[100] Gut microbiota can affect drug metabolism via direct and indirect mechanisms.[101] The direct mechanism is mediated by the microbial enzymes that can modify the chemical structure of the administered drugs.[102] Conversely, the indirect pathway is mediated by the microbial metabolites which affect the expression of host metabolizing enzymes such as cytochrome P450.[103][101] The effects of the gut microbiota on the pharmacokinetics and bioavailability of the drug have been investigated a few decades ago.[104][105][106] These effects can be varied; it could activate the inactive drugs such as lovastatin,[107] inactivate the active drug such as digoxin[108] or induce drug toxicity as in irinotecan.[109] Since then, the impacts of the gut microbiota on the pharmacokinetics of many drugs were heavily studied.[110][100]
The human gut microbiota plays a crucial role in modulating the effect of the administered drugs on the human. Directly, gut microbiota can synthesize and release a series of enzymes with the capability to metabolize drugs such as microbial biotransformation of L-dopa by decarboxylase and dehydroxylase enzymes.[102] On the contrary, gut microbiota may also alter the metabolism of the drugs by modulating the host drug metabolism. This mechanism can be mediated by microbial metabolites or by modifying host metabolites which in turn change the expression of host metabolizing enzymes.[103]
A large number of studies have demonstrated the metabolism of over 50 drugs by the gut microbiota.[110][101] For example, lovastatin (a cholesterol-lowering agent) which is a lactone prodrug is partially activated by the human gut microbiota forming active acid hydroxylated metabolites.[107] Conversely, digoxin (a drug used to treat Congestive Heart Failure) is inactivated by a member of the gut microbiota (i.e. Eggerthella lanta).[111] Eggerthella lanta has a cytochrome-encoding operon up-regulated by digoxin and associated with digoxin-inactivation.[111] Gut microbiota can also modulate the efficacy and toxicity of chemotherapeutic agents such as irinotecan.[112] This effect is derived from the microbiome-encoded β-glucuronidase enzymes which recover the active form of the irinotecan causing gastrointestinal toxicity.[113]
Secondary metabolites
[edit]This microbial community in the gut has a huge biochemical capability to produce distinct secondary metabolites that are sometimes produced from the metabolic conversion of dietary foods such as fibers, endogenous biological compounds such as indole or bile acids.[114][115][116] Microbial metabolites especially short chain fatty acids (SCFAs) and secondary bile acids (BAs) play important roles for the human in health and disease states.[117][118][119]
One of the most important bacterial metabolites produced by the gut microbiota is secondary bile acids (BAs).[116] These metabolites are produced by the bacterial biotransformation of the primary bile acids such as cholic acid (CA) and chenodeoxycholic acid (CDCA) into secondary bile acids (BAs) lithocholic acid (LCA) and deoxy cholic acid (DCA) respectively.[120] Primary bile acids which are synthesized by hepatocytes and stored in the gall bladder possess hydrophobic characters. These metabolites are subsequently metabolized by the gut microbiota into secondary metabolites with increased hydrophobicity.[120] Bile salt hydrolases (BSH) which are conserved across gut microbiota phyla such as Bacteroides, Firmicutes, and Actinobacteria responsible for the first step of secondary bile acids metabolism.[120] Secondary bile acids (BAs) such as DCA and LCA have been demonstrated to inhibit both Clostridioides difficile germination and outgrowth.[119]
Dysbiosis
[edit]The gut microbiota is important for maintaining homeostasis in the intestine. Development of intestinal cancer is associated with an imbalance in the natural microflora (dysbiosis).[121] The secondary bile acid deoxycholic acid is associated with alterations of the microbial community that lead to increased intestinal carcinogenesis.[121] Increased exposure of the colon to secondary bile acids resulting from dysbiosis can cause DNA damage, and such damage can produce carcinogenic mutations in cells of the colon.[122] The high density of bacteria in the colon (about 1012 per ml.) that are subject to dysbiosis compared to the relatively low density in the small intestine (about 102 per ml.) may account for the greater than 10-fold higher incidence of cancer in the colon compared to the small intestine.[122]
Gut–brain axis
[edit]The gut microbiota contributes to digestion and immune modulation, as it plays a role in the gut-brain axis, where microbial metabolites such as short-chain fatty acids and neurotransmitters influence brain function and behavior. The gut–brain axis is the biochemical signaling that takes place between the gastrointestinal tract and the central nervous system.[78] That term has been expanded to include the role of the gut flora in the interplay; the term "microbiome––brain axis" is sometimes used to describe paradigms explicitly including the gut flora.[78][123][124] Broadly defined, the gut–brain axis includes the central nervous system, neuroendocrine and neuroimmune systems including the hypothalamic–pituitary–adrenal axis (HPA axis), sympathetic and parasympathetic arms of the autonomic nervous system including the enteric nervous system, the vagus nerve, and the gut microbiota.[78][124] Studies show links between gut dysbiosis and mental health conditions, indicating a complex interaction that impacts mood and cognitive functions.
A systematic review from 2016 examined the preclinical and small human trials that have been conducted with certain commercially available strains of probiotic bacteria and found that among those tested, Bifidobacterium and Lactobacillus genera (B. longum, B. breve, B. infantis, L. helveticus, L. rhamnosus, L. plantarum, and L. casei), had the most potential to be useful for certain central nervous system disorders.[16]
Alterations in microbiota balance
[edit]Effects of antibiotic use
[edit]Altering the numbers of gut bacteria, for example by taking broad-spectrum antibiotics, may affect the host's health and ability to digest food.[125] Antibiotics can cause antibiotic-associated diarrhea by irritating the bowel directly, changing the levels of microbiota, or allowing pathogenic bacteria to grow.[7] Another harmful effect of antibiotics is the increase in numbers of antibiotic-resistant bacteria found after their use, which, when they invade the host, cause illnesses that are difficult to treat with antibiotics.[125]
Changing the numbers and species of gut microbiota can reduce the body's ability to ferment carbohydrates and metabolize bile acids and may cause diarrhea. Carbohydrates that are not broken down may absorb too much water and cause runny stools, or lack of SCFAs produced by gut microbiota could cause diarrhea.[7]
A reduction in levels of native bacterial species also disrupts their ability to inhibit the growth of harmful species such as C. difficile and Salmonella Kedougou, and these species can get out of hand, though their overgrowth may be incidental and not be the true cause of diarrhea.[6][7][125] Emerging treatment protocols for C. difficile infections involve fecal microbiota transplantation of donor feces (see Fecal transplant).[126] Initial reports of treatment describe success rates of 90%, with few side effects. Efficacy is speculated to result from restoring bacterial balances of bacteroides and firmicutes classes of bacteria.[127]
The composition of the gut microbiome also changes in severe illnesses, due not only to antibiotic use but also to such factors as ischemia of the gut, failure to eat, and immune compromise. Negative effects from this have led to interest in selective digestive tract decontamination, a treatment to kill only pathogenic bacteria and allow the re-establishment of healthy ones.[128]
Antibiotics alter the population of the microbiota in the gastrointestinal tract, and this may change the intra-community metabolic interactions, modify caloric intake by using carbohydrates, and globally affect host metabolic, hormonal, and immune homeostasis.[129]
There is reasonable evidence that taking probiotics containing Lactobacillus species may help prevent antibiotic-associated diarrhea and that taking probiotics with Saccharomyces (e.g., Saccharomyces boulardii ) may help to prevent Clostridioides difficile infection following systemic antibiotic treatment.[130]
Pregnancy
[edit]The gut microbiota of a woman changes as pregnancy advances, with the changes similar to those seen in metabolic syndromes such as diabetes. The change in gut microbiota causes no ill effects. The newborn's gut microbiota resemble the mother's first-trimester samples. The diversity of the microbiome decreases from the first to third trimester, as the numbers of certain species go up.[66][131]
Probiotics, prebiotics, synbiotics, and pharmabiotics
[edit]Probiotics contain live microorganisms. When consumed, they are believed to provide health benefits by altering the microbiome composition.[132][133][134] Current research explores using probiotics as a way to restore the microbial balance of the intestine by stimulating the immune system and inhibiting pro-inflammatory cytokines.[132]
With regard to gut microbiota, prebiotics are typically non-digestible, fiber compounds that pass undigested through the upper part of the gastrointestinal tract and stimulate the growth or activity of advantageous gut flora by acting as substrate for them.[47][135]
Synbiotics refers to food ingredients or dietary supplements combining probiotics and prebiotics in a form of synergism.[136]
The term "pharmabiotics" is used in various ways, to mean: pharmaceutical formulations (standardized manufacturing that can obtain regulatory approval as a drug) of probiotics, prebiotics, or synbiotics;[137] probiotics that have been genetically engineered or otherwise optimized for best performance (shelf life, survival in the digestive tract, etc.);[138] and the natural products of gut flora metabolism (vitamins, etc.).[139]
There is some evidence that treatment with some probiotic strains of bacteria may be effective in irritable bowel syndrome,[140][141] abdominal bloating [142]and chronic idiopathic constipation. Those organisms most likely to result in a decrease of symptoms have included:
- Bifidobacterium breve
- Bifidobacterium infantis
- Enterococcus faecium
- Lactobacillus plantarum
- Lactobacillus reuteri
- Lactobacillus rhamnosus
- Lactobacillus salivarius
- Propionibacterium freudenreichii
- Saccharomyces boulardii
- Escherichia coli Nissle 1917
- Streptococcus thermophilus[143][144][145]
Fecal floatation
[edit]Feces of about 10–15% of people consistently floats in toilet water ('floaters'), while the rest produce feces that sinks ('sinkers') and production of gas causes feces to float.[146] While conventional mice often produce 'floaters', gnotobiotic germfree mice no gut microbiota (bred in germfree isolator) produce 'sinkers', and gut microbiota colonization in germfree mice leads to food transformation to microbial biomass and enrichment of multiple gasogenic bacterial species that turns the 'sinkers' into 'floaters'.[147]
Research
[edit]Tests for whether non-antibiotic drugs may impact human gut-associated bacteria were performed by in vitro analysis on more than 1000 marketed drugs against 40 gut bacterial strains, demonstrating that 24% of the drugs inhibited the growth of at least one of the bacterial strains.[148]
Effects of exercise
[edit]Gut microbiota and exercise have recently been shown to be interconnected. Both moderate and intense exercise are typically part of the training regimen of endurance athletes, but they exert different effects on health. The interconnection between gut microbiota and endurance sports depends upon exercise intensity and training status.[149]
Role in disease
[edit]Bacteria in the digestive tract can contribute to and be affected by disease in various ways. The presence or overabundance of some kinds of bacteria may contribute to inflammatory disorders such as inflammatory bowel disease.[6] Additionally, metabolites from certain members of the gut flora may influence host signalling pathways, contributing to disorders such as obesity and colon cancer.[6] Some gut bacteria may also cause infections and sepsis, for example when they are allowed to pass from the gut into the rest of the body.[6]
Ulcers
[edit]Helicobacter pylori infection can initiate formation of stomach ulcers when the bacteria penetrate the stomach epithelial lining, then causing an inflammatory phagocytotic response.[150] In turn, the inflammation damages parietal cells which release excessive hydrochloric acid into the stomach and produce less of the protective mucus.[151] Injury to the stomach lining, leading to ulcers, develops when gastric acid overwhelms the defensive properties of cells and inhibits endogenous prostaglandin synthesis, reduces mucus and bicarbonate secretion, reduces mucosal blood flow, and lowers resistance to injury.[151] Reduced protective properties of the stomach lining increase vulnerability to further injury and ulcer formation by stomach acid, pepsin, and bile salts.[150][151]
Bowel perforation
[edit]Normally-commensal bacteria can harm the host if they extrude from the intestinal tract.[12][13] Translocation, which occurs when bacteria leave the gut through its mucosal lining, can occur in a number of different diseases.[13] If the gut is perforated, bacteria invade the interstitium, causing a potentially fatal infection.[5]: 715
Inflammatory bowel diseases
[edit]The two main types of inflammatory bowel diseases, Crohn's disease and ulcerative colitis, are chronic inflammatory disorders of the gut; the causes of these diseases are unknown and issues with the gut flora and its relationship with the host have been implicated in these conditions.[15][152][153][154] Additionally, it appears that interactions of gut flora with the gut–brain axis have a role in IBD, with physiological stress mediated through the hypothalamic–pituitary–adrenal axis driving changes to intestinal epithelium and the gut flora in turn releasing factors and metabolites that trigger signaling in the enteric nervous system and the vagus nerve.[4]
The diversity of gut flora appears to be significantly diminished in people with inflammatory bowel diseases compared to healthy people; additionally, in people with ulcerative colitis, Proteobacteria and Actinobacteria appear to dominate; in people with Crohn's, Enterococcus faecium and several Proteobacteria appear to be over-represented.[4]
There is reasonable evidence that correcting gut flora imbalances by taking probiotics with Lactobacilli and Bifidobacteria can reduce visceral pain and gut inflammation in IBD.[130]
Irritable bowel syndrome
[edit]Irritable bowel syndrome is a result of stress and chronic activation of the HPA axis; its symptoms include abdominal pain, changes in bowel movements, and an increase in proinflammatory cytokines. Overall, studies have found that the luminal and mucosal microbiota are changed in irritable bowel syndrome individuals, and these changes can relate to the type of irritation such as diarrhea or constipation. Also, there is a decrease in the diversity of the microbiome with low levels of fecal Lactobacilli and Bifidobacteria, high levels of facultative anaerobic bacteria such as Escherichia coli, and increased ratios of Firmicutes: Bacteroidetes.[124]
Asthma
[edit]With asthma, two hypotheses have been posed to explain its rising prevalence in the developed world. The hygiene hypothesis posits that children in the developed world are not exposed to enough microbes and thus may contain lower prevalence of specific bacterial taxa that play protective roles.[155] The second hypothesis focuses on the Western pattern diet, which lacks whole grains and fiber and has an overabundance of simple sugars.[15] Both hypotheses converge on the role of short-chain fatty acids (SCFAs) in immunomodulation. These bacterial fermentation metabolites are involved in immune signalling that prevents the triggering of asthma and lower SCFA levels are associated with the disease.[155][156] Lacking protective genera such as Lachnospira, Veillonella, Rothia and Faecalibacterium has been linked to reduced SCFA levels.[155] Further, SCFAs are the product of bacterial fermentation of fiber, which is low in the Western pattern diet.[15][156] SCFAs offer a link between gut flora and immune disorders, and as of 2016, this was an active area of research.[15] Similar hypotheses have also been posited for the rise of food and other allergies.[157]
Diabetes mellitus type 1
[edit]The connection between the gut microbiota and diabetes mellitus type 1 has also been linked to SCFAs, such as butyrate and acetate. Diets yielding butyrate and acetate from bacterial fermentation show increased Treg expression.[158] Treg cells downregulate effector T cells, which in turn reduces the inflammatory response in the gut.[159] Butyrate is an energy source for colon cells. butyrate-yielding diets thus decrease gut permeability by providing sufficient energy for the formation of tight junctions.[160] Additionally, butyrate has also been shown to decrease insulin resistance, suggesting gut communities low in butyrate-producing microbes may increase chances of acquiring diabetes mellitus type 2.[161] Butyrate-yielding diets may also have potential colorectal cancer suppression effects.[160]
Obesity and metabolic syndrome
[edit]The gut flora have been implicated in obesity and metabolic syndrome due to a key role in the digestive process; the Western pattern diet appears to drive and maintain changes in the gut flora that in turn change how much energy is derived from food and how that energy is used.[154][162] One aspect of a healthy diet that is often lacking in the Western-pattern diet is fiber and other complex carbohydrates that a healthy gut flora require flourishing; changes to gut flora in response to a Western-pattern diet appear to increase the amount of energy generated by the gut flora which may contribute to obesity and metabolic syndrome.[130] There is also evidence that microbiota influence eating behaviours based on the preferences of the microbiota, which can lead to the host consuming more food eventually resulting in obesity. It has generally been observed that with higher gut microbiome diversity, the microbiota will spend energy and resources on competing with other microbiota and less on manipulating the host. The opposite is seen with lower gut microbiome diversity, and these microbiotas may work together to create host food cravings.[58]
Additionally, the liver plays a dominant role in blood glucose homeostasis by maintaining a balance between the uptake and storage of glucose through the metabolic pathways of glycogenesis and gluconeogenesis. Intestinal lipids regulate glucose homeostasis involving a gut–brain–liver axis. The direct administration of lipids into the upper intestine increases the long chain fatty acyl-coenzyme A (LCFA-CoA) levels in the upper intestines and suppresses glucose production even under subdiaphragmatic vagotomy or gut vagal deafferentation. This interrupts the neural connection between the brain and the gut and blocks the upper intestinal lipids' ability to inhibit glucose production. The gut–brain–liver axis and gut microbiota composition can regulate the glucose homeostasis in the liver and provide potential therapeutic methods to treat obesity and diabetes.[163]
Just as gut flora can function in a feedback loop that can drive the development of obesity, there is evidence that restricting intake of calories (i.e., dieting) can drive changes to the composition of the gut flora.[154]
Other animals
[edit]The composition of the human gut microbiome is similar to that of the other great apes. However, humans' gut biota has decreased in diversity and changed in composition since our evolutionary split from Pan.[164] Humans display increases in Bacteroidetes, a bacterial phylum associated with diets high in animal protein and fat, and decreases in Methanobrevibacter and Fibrobacter, groups that ferment complex plant polysaccharides.[164] These changes are the result of the combined dietary, genetic, and cultural changes humans have undergone since evolutionary divergence from Pan.[citation needed]
In addition to humans and vertebrates, some insects also have complex and diverse gut microbiota that play key nutritional roles.[2] Microbial communities associated with termites can constitute a majority of the weight of the individuals and perform important roles in the digestion of lignocellulose and nitrogen fixation.[165] It is known that the disruption of gut microbiota of termites using agents like antibiotics[166] or boric acid[167] (a common agent used in preventative treatment) causes severe damage to digestive function and leads to the rise of opportunistic pathogens.[167] These communities are host-specific, and closely related insect species share comparable similarities in gut microbiota composition.[168][169] In cockroaches, gut microbiota have been shown to assemble in a deterministic fashion, irrespective of the inoculum;[170] the reason for this host-specific assembly remains unclear. Bacterial communities associated with insects like termites and cockroaches are determined by a combination of forces, primarily diet, but there is some indication that host phylogeny may also be playing a role in the selection of lineages.[168][169]
For more than 51 years it has been known that the administration of low doses of antibacterial agents promotes the growth of farm animals to increase weight gain.[129]
In a study carried out on mice the ratio of Firmicutes and Lachnospiraceae was significantly elevated in animals treated with subtherapeutic doses of different antibiotics. By analyzing the caloric content of faeces and the concentration of small chain fatty acids (SCFAs) in the GI tract, it was concluded that the changes in the composition of microbiota lead to an increased capacity to extract calories from otherwise indigestible constituents, and to an increased production of SCFAs. These findings provide evidence that antibiotics perturb not only the composition of the GI microbiome but also its metabolic capabilities, specifically with respect to SCFAs.[129]
See also
[edit]- Colonisation resistance
- List of human flora
- List of microbiota species of the lower reproductive tract of women
- Skin flora
- Verotoxin-producing Escherichia coli
Notes
[edit]References
[edit]- ^ Moszak, M; Szulińska, M; Bogdański, P (15 April 2020). "You Are What You Eat – The Relationship between Diet, Microbiota, and Metabolic Disorders-A Review". Nutrients. 12 (4): 1096. doi:10.3390/nu12041096. PMC 7230850. PMID 32326604.
- ^ a b Engel, P.; Moran, N. (2013). "The gut microbiota of insects–diversity in structure and function". FEMS Microbiology Reviews. 37 (5): 699–735. doi:10.1111/1574-6976.12025. PMID 23692388.
- ^ Segata, N; Boernigen, D; Tickle, TL; Morgan, XC; Garrett, WS; Huttenhower, C (14 May 2013). "Computational meta'omics for microbial community studies". Molecular Systems Biology. 9: 666. doi:10.1038/msb.2013.22. PMC 4039370. PMID 23670539.
- ^ a b c d Saxena, R.; Sharma, V.K (2016). "A Metagenomic Insight Into the Human Microbiome: Its Implications in Health and Disease". In Kumar, D.; S. Antonarakis (eds.). Medical and Health Genomics. Elsevier Science. p. 117. doi:10.1016/B978-0-12-420196-5.00009-5. ISBN 978-0-12-799922-7.
- ^ a b c d e f g h i j k Sherwood, Linda; Willey, Joanne; Woolverton, Christopher J. (2013). Prescott's Microbiology. McGraw-Hill Education. pp. 713–721. ISBN 978-0-07-340240-6.
- ^ a b c d e f g h i j k l m n o Guarner, F; Malagelada, J (2003). "Gut flora in health and disease". The Lancet. 361 (9356): 512–519. doi:10.1016/S0140-6736(03)12489-0. PMID 12583961.
- ^ a b c d e f g h i j k Beaugerie, Laurent; Petit, Jean-Claude (2004). "Antibiotic-associated diarrhoea". Best Practice & Research Clinical Gastroenterology. 18 (2): 337–352. doi:10.1016/j.bpg.2003.10.002. PMID 15123074.
- ^ a b Stephen, A. M.; Cummings, J. H. (1980). "The Microbial Contribution to Human Faecal Mass". Journal of Medical Microbiology. 13 (1): 45–56. doi:10.1099/00222615-13-1-45. PMID 7359576.
- ^ a b c d e Quigley, E. M (2013). "Gut bacteria in health and disease". Gastroenterology & Hepatology. 9 (9): 560–569. PMC 3983973. PMID 24729765.
- ^ Turnbaugh, Peter J.; Ley, Ruth E.; Hamady, Micah; Fraser-Liggett, Claire M.; Knight, Rob; Gordon, Jeffrey I. (October 2007). "The Human Microbiome Project". Nature. 449 (7164): 804–810. Bibcode:2007Natur.449..804T. doi:10.1038/nature06244. PMC 3709439. PMID 17943116.
- ^ Ma, Guangyu; Shi, Yuguo; Meng, Lulu; Fan, Haolong; Tang, Xiaomei; Luo, Huijuan; Wang, Dongju; Zhou, Juan; Xiao, Xiaomin (2023). "Factors affecting the early establishment of neonatal intestinal flora and its intervention measures". Frontiers in Cellular and Infection Microbiology. 13. doi:10.3389/fcimb.2023.1295111. PMC 10722192. PMID 38106467.
- ^ a b c d e f g h i j k l m n o p Sommer, Felix; Bäckhed, Fredrik (2013). "The gut microbiota – masters of host development and physiology". Nature Reviews Microbiology. 11 (4): 227–238. doi:10.1038/nrmicro2974. PMID 23435359.
- ^ a b c d Faderl, Martin; Noti, Mario; Corazza, Nadia; Mueller, Christoph (2015). "Keeping bugs in check: The mucus layer as a critical component in maintaining intestinal homeostasis". IUBMB Life. 67 (4): 275–285. doi:10.1002/iub.1374. PMID 25914114.
- ^ a b c d e f Clarke, Gerard; Stilling, Roman M; Kennedy, Paul J; Stanton, Catherine; Cryan, John F; Dinan, Timothy G (2014). "Minireview: Gut Microbiota: The Neglected Endocrine Organ". Molecular Endocrinology. 28 (8): 1221–1238. doi:10.1210/me.2014-1108. PMC 5414803. PMID 24892638.
- ^ a b c d e f Shen, Sj; Wong, Connie HY (2016). "Bugging inflammation: Role of the gut microbiota". Clinical & Translational Immunology. 5 (4): e72. doi:10.1038/cti.2016.12. PMC 4855262. PMID 27195115.
- ^ a b Wang, Huiying; Lee, In-Seon; Braun, Christoph; Enck, Paul (2016). "Effect of Probiotics on Central Nervous System Functions in Animals and Humans: A Systematic Review". Journal of Neurogastroenterology and Motility. 22 (4): 589–605. doi:10.5056/jnm16018. PMC 5056568. PMID 27413138.
- ^ Salvadori, M (20 March 2024). "Update on the gut microbiome in health and diseases". World J Methodol. 14 (1). doi:10.5662/wjm.v14.i1.89196. PMC 10989414. PMID 38577200.
- ^ a b c d e f Sears, Cynthia L. (2005). "A dynamic partnership: Celebrating our gut flora". Anaerobe. 11 (5): 247–251. doi:10.1016/j.anaerobe.2005.05.001. PMID 16701579.
- ^ Shapira, Michael (July 2016). "Gut Microbiotas and Host Evolution: Scaling Up Symbiosis". Trends in Ecology & Evolution. 31 (7): 539–549. Bibcode:2016TEcoE..31..539S. doi:10.1016/j.tree.2016.03.006. PMID 27039196.
- ^ Walker, Alan W.; Hoyles, Lesley (August 2023). "Human microbiome myths and misconceptions". Nature Microbiology. 8 (8): 1392–1396. doi:10.1038/s41564-023-01426-7. PMID 37524974.
- ^ Lozupone, Catherine A.; Stombaugh, Jesse I.; Gordon, Jeffrey I.; Jansson, Janet K.; Knight, Rob (2012). "Diversity, stability and resilience of the human gut microbiota". Nature. 489 (7415): 220–230. Bibcode:2012Natur.489..220L. doi:10.1038/nature11550. PMC 3577372. PMID 22972295.
- ^ Qin, Junjie; Li, Ruiqiang; Raes, Jeroen; Arumugam, Manimozhiyan; Burgdorf, Kristoffer Solvsten; Manichanh, Chaysavanh; Nielsen, Trine; Pons, Nicolas; Levenez, Florence; Yamada, Takuji; Mende, Daniel R.; Li, Junhua; Xu, Junming; Li, Shaochuan; Li, Dongfang; Cao, Jianjun; Wang, Bo; Liang, Huiqing; Zheng, Huisong; Xie, Yinlong; Tap, Julien; Lepage, Patricia; Bertalan, Marcelo; Batto, Jean-Michel; Hansen, Torben; Le Paslier, Denis; Linneberg, Allan; Nielsen, H. Bjørn; Pelletier, Eric; Renault, Pierre (2010). "A human gut microbial gene catalogue established by metagenomic sequencing". Nature. 464 (7285): 59–65. Bibcode:2010Natur.464...59.. doi:10.1038/nature08821. PMC 3779803. PMID 20203603.
- ^ Shanahan, Fergus (2002). "The host–microbe interface within the gut". Best Practice & Research Clinical Gastroenterology. 16 (6): 915–931. doi:10.1053/bega.2002.0342. PMID 12473298.
- ^ Tap, Julien; Mondot, Stanislas; Levenez, Florence; Pelletier, Eric; Caron, Christophe; Furet, Jean-Pierre; Ugarte, Edgardo; Muñoz-Tamayo, Rafael; Paslier, Denis L. E.; Nalin, Renaud; Dore, Joel; Leclerc, Marion (2009). "Towards the human intestinal microbiota phylogenetic core". Environmental Microbiology. 11 (10): 2574–2584. Bibcode:2009EnvMi..11.2574T. doi:10.1111/j.1462-2920.2009.01982.x. PMID 19601958.
- ^ a b O'Hara, Ann M; Shanahan, Fergus (2006). "The gut flora as a forgotten organ". EMBO Reports. 7 (7): 688–693. doi:10.1038/sj.embor.7400731. PMC 1500832. PMID 16819463.
- ^ Khanna, Sahil; Tosh, Pritish K (2014). "A Clinician's Primer on the Role of the Microbiome in Human Health and Disease". Mayo Clinic Proceedings. 89 (1): 107–114. doi:10.1016/j.mayocp.2013.10.011. PMID 24388028.
- ^ a b Cui, Lijia; Morris, Alison; Ghedin, Elodie (2013). "The human mycobiome in health and disease". Genome Medicine. 5 (7): 63. doi:10.1186/gm467. PMC 3978422. PMID 23899327.
- ^ Erdogan, Askin; Rao, Satish S. C (2015). "Small Intestinal Fungal Overgrowth". Current Gastroenterology Reports. 17 (4): 16. doi:10.1007/s11894-015-0436-2. PMID 25786900.
- ^ Hallen-Adams, Heather E.; Suhr, Mallory J. (3 April 2017). "Fungi in the healthy human gastrointestinal tract". Virulence. 8 (3): 352–358. doi:10.1080/21505594.2016.1247140. PMC 5411236. PMID 27736307.
- ^ a b c "What fungi live in the gut? Meet the gut mycobiome". Quadram Institute. Retrieved 2024-07-25.
- ^ Klein, Alice (20 October 2019). "Man's body brews its own beer after yeast take over his gut microbiome". New Scientist. Retrieved 2024-07-25.
- ^ Painter, Kelly; Cordell, Barbara J.; Sticco, Kristin L. (2024), "Auto-Brewery Syndrome", StatPearls, Treasure Island (FL): StatPearls Publishing, PMID 30020718, retrieved 2024-07-25
- ^ Browne, Hilary P; Shao, Yan; Lawley, Trevor D (October 2022). "Mother–infant transmission of human microbiota". Current Opinion in Microbiology. 69: 102173. doi:10.1016/j.mib.2022.102173. PMID 35785616.
- ^ Bello, Maria G. Dominguez; Knight, Rob; Gilbert, Jack A.; Blaser, Martin J. (4 October 2018). "Preserving microbial diversity". Science. 362 (6410): 33–34. Bibcode:2018Sci...362...33B. doi:10.1126/science.aau8816. PMID 30287652.
- ^ a b Arumugam, Manimozhiyan; Raes, Jeroen; Pelletier, Eric; Le Paslier, Denis; Yamada, Takuji; Mende, Daniel R.; Fernandes, Gabriel R.; Tap, Julien; Bruls, Thomas; Batto, Jean-Michel; Bertalan, Marcelo; Borruel, Natalia; Casellas, Francesc; Fernandez, Leyden; Gautier, Laurent; Hansen, Torben; Hattori, Masahira; Hayashi, Tetsuya; Kleerebezem, Michiel; Kurokawa, Ken; Leclerc, Marion; Levenez, Florence; Manichanh, Chaysavanh; Nielsen, H. Bjørn; Nielsen, Trine; Pons, Nicolas; Poulain, Julie; Qin, Junjie; Sicheritz-Ponten, Thomas; Tims, Sebastian (2011). "Enterotypes of the human gut microbiome". Nature. 473 (7346): 174–180. Bibcode:2011Natur.473..174.. doi:10.1038/nature09944. PMC 3728647. PMID 21508958.
- ^ Wu, G. D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S. A.; Bewtra, M.; Knights, D.; Walters, W. A.; Knight, R.; Sinha, R.; Gilroy, E.; Gupta, K.; Baldassano, R.; Nessel, L.; Li, H.; Bushman, F. D.; Lewis, J. D. (2011). "Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes". Science. 334 (6052): 105–108. Bibcode:2011Sci...334..105W. doi:10.1126/science.1208344. PMC 3368382. PMID 21885731.
- ^ Zimmer, Carl (April 20, 2011). "Bacteria Divide People Into 3 Types, Scientists Say". The New York Times. Retrieved April 21, 2011.
a group of scientists now report just three distinct ecosystems in the guts of people they have studied.
- ^ Knights, Dan; Ward, Tonya; McKinlay, Christopher; Miller, Hannah; Gonzalez, Antonio; McDonald, Daniel; Knight, Rob (8 October 2014). "Rethinking "Enterotypes"". Cell Host & Microbe. 16 (4): 433–437. doi:10.1016/j.chom.2014.09.013. PMC 5558460. PMID 25299329.
- ^ a b Nardone, G; Compare, D (June 2015). "The human gastric microbiota: Is it time to rethink the pathogenesis of stomach diseases?". United European Gastroenterology Journal. 3 (3): 255–60. doi:10.1177/2050640614566846. PMC 4480535. PMID 26137299.
- ^ Yao, X; Smolka, AJ (June 2019). "Gastric Parietal Cell Physiology and Helicobacter pylori-Induced Disease". Gastroenterology. 156 (8): 2158–2173. doi:10.1053/j.gastro.2019.02.036. PMC 6715393. PMID 30831083.
- ^ Todar, Kenneth (2012). "The Normal Bacterial Flora of Humans". Todar's Online Textbook of Bacteriology. Retrieved June 25, 2016.
- ^ Quigley, Eamonn M.M; Quera, Rodrigo (2006). "Small Intestinal Bacterial Overgrowth: Roles of Antibiotics, Prebiotics, and Probiotics". Gastroenterology. 130 (2): S78–90. doi:10.1053/j.gastro.2005.11.046. PMID 16473077.
- ^ Adams, M. R.; Moss, M. O. (2007). Food Microbiology. doi:10.1039/9781847557940. ISBN 978-0-85404-284-5.[page needed]
- ^ a b "The normal gut flora" (PDF) (slideshow). 2004. Archived from the original (PDF) on 2004-05-26. Retrieved 2023-01-02 – via University of Glasgow.
- ^ Braune A, Blaut M (2016). "Bacterial species involved in the conversion of dietary flavonoids in the human gut". Gut Microbes. 7 (3): 216–234. doi:10.1080/19490976.2016.1158395. PMC 4939924. PMID 26963713.
- ^ a b Steinhoff, U (2005). "Who controls the crowd? New findings and old questions about the intestinal microflora". Immunology Letters. 99 (1): 12–16. doi:10.1016/j.imlet.2004.12.013. PMID 15894105.
- ^ a b c d e f Gibson, Glenn R (2004). "Fibre and effects on probiotics (the prebiotic concept)". Clinical Nutrition Supplements. 1 (2): 25–31. doi:10.1016/j.clnu.2004.09.005.
- ^ Miquel, S; Martín, R; Rossi, O; Bermúdez-Humarán, LG; Chatel, JM; Sokol, H; Thomas, M; Wells, JM; Langella, P (2013). "Faecalibacterium prausnitzii and human intestinal health". Current Opinion in Microbiology. 16 (3): 255–261. doi:10.1016/j.mib.2013.06.003. PMID 23831042.
- ^ Ley, Ruth E (2010). "Obesity and the human microbiome". Current Opinion in Gastroenterology. 26 (1): 5–11. doi:10.1097/MOG.0b013e328333d751. PMID 19901833.
- ^ Nash, Andrea K; Auchtung, Thomas A; Wong, Matthew C; Smith, Daniel P; Gesell, Jonathan R; Ross, Matthew C; Stewart, Christopher J; Metcalf, Ginger A; Muzny, Donna M; Gibbs, Richard A; Ajami, Nadim J; Petrosino, Joseph F (2017). "The gut mycobiome of the Human Microbiome Project healthy cohort". Microbiome. 5 (1): 153. doi:10.1186/s40168-017-0373-4. PMC 5702186. PMID 29178920.
- ^ Scarpellini, Emidio; Ianiro, Gianluca; Attili, Fabia; Bassanelli, Chiara; De Santis, Adriano; Gasbarrini, Antonio (2015). "The human gut microbiota and virome: Potential therapeutic implications". Digestive and Liver Disease. 47 (12): 1007–1012. doi:10.1016/j.dld.2015.07.008. PMC 7185617. PMID 26257129.
- ^ Gerritsen, Jacoline; Smidt, Hauke; Rijkers, Ger; de Vos, Willem (27 May 2011). "Intestinal microbiota in human health and disease: the impact of probiotics". Genes & Nutrition. 6 (3): 209–240. doi:10.1007/s12263-011-0229-7. PMC 3145058. PMID 21617937.
- ^ a b c d e f Yatsunenko, T.; Rey, F. E.; Manary, M. J.; Trehan, I.; Dominguez-Bello, M. G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R. N.; Anokhin, A. P.; Heath, A. C.; Warner, B.; Reeder, J.; Kuczynski, J.; Caporaso, J. G.; Lozupone, C. A.; Lauber, C.; Clemente, J. C.; Knights, D.; Knight, R.; Gordon, J. I. (2012). "Human gut microbiome viewed across age and geography". Nature. 486 (7402): 222–227. Bibcode:2012Natur.486..222Y. doi:10.1038/nature11053. PMC 3376388. PMID 22699611.
- ^ De Filippo, C; Cavalieri, D; Di Paola, M; Ramazzotti, M; Poullet, J. B; Massart, S; Collini, S; Pieraccini, G; Lionetti, P (2010). "Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa". Proceedings of the National Academy of Sciences. 107 (33): 14691–14696. Bibcode:2010PNAS..10714691D. doi:10.1073/pnas.1005963107. PMC 2930426. PMID 20679230.
- ^ Jonkers, Daisy M.A.E. (2016). "Microbial perturbations and modulation in conditions associated with malnutrition and malabsorption". Best Practice & Research Clinical Gastroenterology. 30 (2): 161–172. doi:10.1016/j.bpg.2016.02.006. PMID 27086883.
- ^ Million, Matthieu; Diallo, Aldiouma; Raoult, Didier (May 2017). "Gut microbiota and malnutrition" (PDF). Microbial Pathogenesis. 106: 127–138. doi:10.1016/j.micpath.2016.02.003. PMID 26853753.
- ^ Rytter, Maren Johanne Heilskov; Kolte, Lilian; Briend, André; Friis, Henrik; Christensen, Vibeke Brix (2014). "The Immune System in Children with Malnutrition – A Systematic Review". PLOS ONE. 9 (8): e105017. Bibcode:2014PLoSO...9j5017R. doi:10.1371/journal.pone.0105017. PMC 4143239. PMID 25153531.
- ^ a b Alcock, Joe; Maley, Carlo C; Aktipis, C. Athena (2014). "Is eating behavior manipulated by the gastrointestinal microbiota? Evolutionary pressures and potential mechanisms". BioEssays. 36 (10): 940–949. doi:10.1002/bies.201400071. PMC 4270213. PMID 25103109.
- ^ a b c Renson, Audrey; Herd, Pamela; Dowd, Jennifer B. (2020). "Sick Individuals and Sick (Microbial) Populations: Challenges in Epidemiology and the Microbiome". Annual Review of Public Health. 41: 63–80. doi:10.1146/annurev-publhealth-040119-094423. PMC 9713946. PMID 31635533.
- ^ Colella, M., Charitos, I. A., Ballini, A., Cafiero, C., Topi, S., Palmirotta, R., & Santacroce, L. (2023). Microbiota revolution: How gut microbes regulate our lives. World journal of gastroenterology, 29(28), 4368–4383. https://doi.org/10.3748/wjg.v29.i28.4368
- ^ Colella, M., Charitos, I. A., Ballini, A., Cafiero, C., Topi, S., Palmirotta, R., & Santacroce, L. (2023). Microbiota revolution: How gut microbes regulate our lives. World journal of gastroenterology, 29(28), 4368–4383. https://doi.org/10.3748/wjg.v29.i28.4368
- ^ Turroni, Francesca; Peano, Clelia; Pass, Daniel A; Foroni, Elena; Severgnini, Marco; Claesson, Marcus J; Kerr, Colm; Hourihane, Jonathan; Murray, Deirdre; Fuligni, Fabio; Gueimonde, Miguel; Margolles, Abelardo; De Bellis, Gianluca; o'Toole, Paul W; Van Sinderen, Douwe; Marchesi, Julian R; Ventura, Marco (2012). "Diversity of Bifidobacteria within the Infant Gut Microbiota". PLOS ONE. 7 (5): e36957. Bibcode:2012PLoSO...736957T. doi:10.1371/journal.pone.0036957. PMC 3350489. PMID 22606315.
- ^ Davenport, Emily R.; Sanders, Jon G.; Song, Se Jin; Amato, Katherine R.; Clark, Andrew G.; Knight, Rob (December 2017). "The human microbiome in evolution". BMC Biology. 15 (1): 127. doi:10.1186/s12915-017-0454-7. PMC 5744394. PMID 29282061.
- ^ Perez-Muñoz, Maria Elisa; Arrieta, Marie-Claire; Ramer-Tait, Amanda E; Walter, Jens (2017). "A critical assessment of the 'sterile womb' and 'in utero colonization' hypotheses: Implications for research on the pioneer infant microbiome". Microbiome. 5 (1): 48. doi:10.1186/s40168-017-0268-4. PMC 5410102. PMID 28454555.
- ^ Matamoros, Sebastien; Gras-Leguen, Christele; Le Vacon, Françoise; Potel, Gilles; de la Cochetiere, Marie-France (2013). "Development of intestinal microbiota in infants and its impact on health". Trends in Microbiology. 21 (4): 167–173. doi:10.1016/j.tim.2012.12.001. PMID 23332725.
- ^ a b c d e f g Mueller, Noel T.; Bakacs, Elizabeth; Combellick, Joan; Grigoryan, Zoya; Dominguez-Bello, Maria G. (2015). "The infant microbiome development: mom matters". Trends in Molecular Medicine. 21 (2): 109–117. doi:10.1016/j.molmed.2014.12.002. PMC 4464665. PMID 25578246.
- ^ Jiménez, Esther; Fernández, Leonides; Marín, María L; Martín, Rocío; Odriozola, Juan M; Nueno-Palop, Carmen; Narbad, Arjan; Olivares, Mónica; Xaus, Jordi; Rodríguez, Juan M (2005). "Isolation of Commensal Bacteria from Umbilical Cord Blood of Healthy Neonates Born by Cesarean Section". Current Microbiology. 51 (4): 270–274. doi:10.1007/s00284-005-0020-3. PMID 16187156.
- ^ Collado, Maria Carmen; Rautava, Samuli; Aakko, Juhani; Isolauri, Erika; Salminen, Seppo (2016). "Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid". Scientific Reports. 6: 23129. Bibcode:2016NatSR...623129C. doi:10.1038/srep23129. PMC 4802384. PMID 27001291.
- ^ Jiménez, Esther; Marín, María L.; Martín, Rocío; Odriozola, Juan M.; Olivares, Mónica; Xaus, Jordi; Fernández, Leonides; Rodríguez, Juan M. (2008). "Is meconium from healthy newborns actually sterile?". Research in Microbiology. 159 (3): 187–193. doi:10.1016/j.resmic.2007.12.007. PMID 18281199.
- ^ Perez-Muñoz, Maria Elisa; Arrieta, Marie-Claire; Ramer-Tait, Amanda E; Walter, Jens (2017). "A critical assessment of the "sterile womb" and "in utero colonization" hypotheses: Implications for research on the pioneer infant microbiome". Microbiome. 5 (1): 48. doi:10.1186/s40168-017-0268-4. PMC 5410102. PMID 28454555.
- ^ Adlerberth, I; Wold, AE (2009). "Establishment of the gut microbiota in Western infants". Acta Paediatrica. 98 (2): 229–238. doi:10.1111/j.1651-2227.2008.01060.x. PMID 19143664.
- ^ "Babies' gut bacteria affected by delivery method". ScienceDaily (Press release). Wellcome Trust Sanger Institute. 18 September 2019. Archived from the original on 24 November 2021.
- ^ Coppa, G. V.; Zampini, L.; Galeazzi, T.; Gabrielli, O. (2006). "Prebiotics in human milk: A review". Digestive and Liver Disease. 38: S291–294. doi:10.1016/S1590-8658(07)60013-9. PMID 17259094.
- ^ Mady, Eman A.; Doghish, Ahmed S.; El-Dakroury, Walaa A.; Elkhawaga, Samy Y.; Ismail, Ahmed; El-Mahdy, Hesham A.; Elsakka, Elsayed G.E.; El-Husseiny, Hussein M. (July 2023). "Impact of the mother's gut microbiota on infant microbiome and brain development". Neuroscience & Biobehavioral Reviews. 150: 105195. doi:10.1016/j.neubiorev.2023.105195. PMID 37100161.
- ^ Fanaro, S; Chierici, R; Guerrini, P; Vigi, V (2007). "Intestinal microflora in early infancy: Composition and development". Acta Paediatrica. 92 (441): 48–55. doi:10.1111/j.1651-2227.2003.tb00646.x. PMID 14599042.
- ^ Yassour, Moran; Vatanen, Tommi; Siljander, Heli; Hämäläinen, Anu-Maaria; Härkönen, Taina; Ryhänen, Samppa J; Franzosa, Eric A; Vlamakis, Hera; Huttenhower, Curtis; Gevers, Dirk; Lander, Eric S; Knip, Mikael; Xavier, Ramnik J (2016). "Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability". Science Translational Medicine. 8 (343): 343ra81. doi:10.1126/scitranslmed.aad0917. PMC 5032909. PMID 27306663.
- ^ Gibson, G. R.; Roberfroid, M. B. (1995). "Dietary modulation of the human colonic microbiota: Introducing the concept of prebiotics". The Journal of Nutrition. 125 (6): 1401–1412. doi:10.1093/jn/125.6.1401. PMID 7782892.
- ^ a b c d Wang, Yan; Kasper, Lloyd H (2014). "The role of microbiome in central nervous system disorders". Brain, Behavior, and Immunity. 38: 1–12. doi:10.1016/j.bbi.2013.12.015. PMC 4062078. PMID 24370461.
- ^ a b Yoon, My Young; Lee, Keehoon; Yoon, Sang Sun (2014). "Protective role of gut commensal microbes against intestinal infections". Journal of Microbiology. 52 (12): 983–989. doi:10.1007/s12275-014-4655-2. PMID 25467115.
- ^ Reinoso Webb, Cynthia; Koboziev, Iurii; Furr, Kathryn L; Grisham, Matthew B (2016). "Protective and pro-inflammatory roles of intestinal bacteria". Pathophysiology. 23 (2): 67–80. doi:10.1016/j.pathophys.2016.02.002. PMC 4867289. PMID 26947707.
- ^ Mantis, N J; Rol, N; Corthésy, B (2011). "Secretory IgA's complex roles in immunity and mucosal homeostasis in the gut". Mucosal Immunology. 4 (6): 603–611. doi:10.1038/mi.2011.41. PMC 3774538. PMID 21975936.
- ^ a b Peterson, Lance W; Artis, David (2014). "Intestinal epithelial cells: Regulators of barrier function and immune homeostasis". Nature Reviews Immunology. 14 (3): 141–153. doi:10.1038/nri3608. PMID 24566914.
- ^ a b Honda, Kenya; Littman, Dan R (2016). "The microbiota in adaptive immune homeostasis and disease". Nature. 535 (7610): 75–84. Bibcode:2016Natur.535...75H. doi:10.1038/nature18848. PMID 27383982.
- ^ a b Levy, M.; Thaiss, C.A.; Elinav, E. (2016). "Metabolites: messengers between the microbiota and the immune system". Genes & Development. 30 (14): 1589–1597. doi:10.1101/gad.284091.116. PMC 4973288. PMID 27474437.
- ^ a b c d e f g h i Zhang LS, Davies SS (April 2016). "Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions". Genome Med. 8 (1): 46. doi:10.1186/s13073-016-0296-x. PMC 4840492. PMID 27102537.
Lactobacillus spp. convert tryptophan to indole-3-aldehyde (I3A) through unidentified enzymes [125]. Clostridium sporogenes convert tryptophan to IPA [6], likely via a tryptophan deaminase. ... IPA also potently scavenges hydroxyl radicals
Table 2: Microbial metabolites: their synthesis, mechanisms of action, and effects on health and disease
Figure 1: Molecular mechanisms of action of indole and its metabolites on host physiology and disease - ^ Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G (March 2009). "Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites". Proc. Natl. Acad. Sci. U.S.A. 106 (10): 3698–3703. Bibcode:2009PNAS..106.3698W. doi:10.1073/pnas.0812874106. PMC 2656143. PMID 19234110.
Production of IPA was shown to be completely dependent on the presence of gut microflora and could be established by colonization with the bacterium Clostridium sporogenes.
IPA metabolism diagram - ^ "3-Indolepropionic acid". Human Metabolome Database. University of Alberta. Retrieved 12 June 2018.
- ^ Chyan YJ, Poeggeler B, Omar RA, Chain DG, Frangione B, Ghiso J, Pappolla MA (July 1999). "Potent neuroprotective properties against the Alzheimer beta-amyloid by an endogenous melatonin-related indole structure, indole-3-propionic acid". J. Biol. Chem. 274 (31): 21937–21942. doi:10.1074/jbc.274.31.21937. PMID 10419516. S2CID 6630247.
[Indole-3-propionic acid (IPA)] has previously been identified in the plasma and cerebrospinal fluid of humans, but its functions are not known. ... In kinetic competition experiments using free radical-trapping agents, the capacity of IPA to scavenge hydroxyl radicals exceeded that of melatonin, an indoleamine considered to be the most potent naturally occurring scavenger of free radicals. In contrast with other antioxidants, IPA was not converted to reactive intermediates with pro-oxidant activity.
- ^ Hopper, Christopher P.; De La Cruz, Ladie Kimberly; Lyles, Kristin V.; Wareham, Lauren K.; Gilbert, Jack A.; Eichenbaum, Zehava; Magierowski, Marcin; Poole, Robert K.; Wollborn, Jakob; Wang, Binghe (23 December 2020). "Role of Carbon Monoxide in Host–Gut Microbiome Communication". Chemical Reviews. 120 (24): 13273–13311. doi:10.1021/acs.chemrev.0c00586. PMID 33089988.
- ^ Rajilić-Stojanović, Mirjana; De Vos, Willem M (2014). "The first 1000 cultured species of the human gastrointestinal microbiota". FEMS Microbiology Reviews. 38 (5): 996–1047. doi:10.1111/1574-6976.12075. PMC 4262072. PMID 24861948.
- ^ Hill, M J (March 1997). "Intestinal flora and endogenous vitamin synthesis". European Journal of Cancer Prevention. 6: S43–S45. doi:10.1097/00008469-199703001-00009. PMID 9167138.
- ^ "The Microbiome". Tufts Now. 2013-09-17. Retrieved 2020-12-09.
- ^ Moraïs, Sarah; Winkler, Sarah; Zorea, Alvah; Levin, Liron; Nagies, Falk S. P.; Kapust, Nils; Lamed, Eva; Artan-Furman, Avital; Bolam, David N.; Yadav, Madhav P.; Bayer, Edward A.; Martin, William F.; Mizrahi, Itzhak (15 March 2024). "Cryptic diversity of cellulose-degrading gut bacteria in industrialized humans". Science. 383 (6688): eadj9223. Bibcode:2024Sci...383j9223M. doi:10.1126/science.adj9223. PMC 7615765. PMID 38484069.
- ^ a b c d e ElRakaiby M, Dutilh BE, Rizkallah MR, Boleij A, Cole JN, Aziz RK (July 2014). "Pharmacomicrobiomics: the impact of human microbiome variations on systems pharmacology and personalized therapeutics". Omics. 18 (7): 402–414. doi:10.1089/omi.2014.0018. PMC 4086029. PMID 24785449.
- ^ a b c Cho I, Blaser MJ (March 2012). "The human microbiome: at the interface of health and disease". Nature Reviews. Genetics. 13 (4): 260–270. doi:10.1038/nrg3182. PMC 3418802. PMID 22411464.
The composition of the microbiome varies by anatomical site (Figure 1). The primary determinant of community composition is anatomical location: interpersonal variation is substantial23,24 and is higher than the temporal variability seen at most sites in a single individual25.
- ^ Hutter T, Gimbert C, Bouchard F, Lapointe FJ (2015). "Being human is a gut feeling". Microbiome. 3: 9. doi:10.1186/s40168-015-0076-7. PMC 4359430. PMID 25774294.
- ^ Kumar, Kundan; Dhoke, Gaurao V.; Sharma, Ashok K.; Jaiswal, Shubham K.; Sharma, Vineet K. (July 2019). "Mechanistic elucidation of amphetamine metabolism by tyramine oxidase from human gut microbiota using molecular dynamics simulations". Journal of Cellular Biochemistry. 120 (7): 11206–11215. doi:10.1002/jcb.28396. PMID 30701587.
- ^ Sousa, Tiago; Paterson, Ronnie; Moore, Vanessa; Carlsson, Anders; Abrahamsson, Bertil; Basit, Abdul W (2008). "The gastrointestinal microbiota as a site for the biotransformation of drugs". International Journal of Pharmaceutics. 363 (1–2): 1–25. doi:10.1016/j.ijpharm.2008.07.009. PMID 18682282.
- ^ Haiser, H. J; Gootenberg, D. B; Chatman, K; Sirasani, G; Balskus, E. P; Turnbaugh, P. J (2013). "Predicting and Manipulating Cardiac Drug Inactivation by the Human Gut Bacterium Eggerthella lenta". Science. 341 (6143): 295–298. Bibcode:2013Sci...341..295H. doi:10.1126/science.1235872. PMC 3736355. PMID 23869020.
- ^ a b Koppel, Nitzan; Maini Rekdal, Vayu; Balskus, Emily P. (23 June 2017). "Chemical transformation of xenobiotics by the human gut microbiota". Science. 356 (6344). doi:10.1126/science.aag2770. PMC 5534341. PMID 28642381.
- ^ a b c Spanogiannopoulos, Peter; Bess, Elizabeth N.; Carmody, Rachel N.; Turnbaugh, Peter J. (May 2016). "The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism". Nature Reviews Microbiology. 14 (5): 273–287. doi:10.1038/nrmicro.2016.17. PMC 5243131. PMID 26972811.
- ^ a b Maini Rekdal, Vayu; Bess, Elizabeth N.; Bisanz, Jordan E.; Turnbaugh, Peter J.; Balskus, Emily P. (14 June 2019). "Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism". Science. 364 (6445). doi:10.1126/science.aau6323. PMC 7745125. PMID 31196984.
- ^ a b Dempsey, Joseph L.; Cui, Julia Yue (December 2019). "Microbiome Is a Functional Modifier of P450 Drug Metabolism". Current Pharmacology Reports. 5 (6): 481–490. doi:10.1007/s40495-019-00200-w. PMC 7731899. PMID 33312848.
- ^ Boerner, Udo; Abbott, Seth; Roe, Robert L. (January 1975). "The Metabolism of Morphine and Heroin in Man". Drug Metabolism Reviews. 4 (1): 39–73. doi:10.3109/03602537508993748. PMID 1204496.
- ^ Dobkin, Jay F.; Saha, Jnan R.; Butler, Vincent P.; Neu, Harold C.; Lindenbaum, John (15 April 1983). "Digoxin-Inactivating Bacteria: Identification in Human Gut Flora". Science. 220 (4594): 325–327. doi:10.1126/science.6836275. PMID 6836275.
- ^ Sahota, S. S.; Bramley, P. M.; Menzies, I. S. (February 1982). "The Fermentation of Lactulose by Colonic Bacteria". Microbiology. 128 (2): 319–325. doi:10.1099/00221287-128-2-319. PMID 6804597.
- ^ a b Yoo, Dae-Hyoung; Kim, In Sook; Van Le, Thi Kim; Jung, Il-Hoon; Yoo, Hye Hyun; Kim, Dong-Hyun (September 2014). "Gut Microbiota-Mediated Drug Interactions between Lovastatin and Antibiotics". Drug Metabolism and Disposition. 42 (9): 1508–1513. doi:10.1124/dmd.114.058354. PMID 24947972.
- ^ Haiser, Henry J.; Gootenberg, David B.; Chatman, Kelly; Sirasani, Gopal; Balskus, Emily P.; Turnbaugh, Peter J. (19 July 2013). "Predicting and Manipulating Cardiac Drug Inactivation by the Human Gut Bacterium Eggerthella lenta". Science. 341 (6143): 295–298. Bibcode:2013Sci...341..295H. doi:10.1126/science.1235872. PMC 3736355. PMID 23869020.
- ^ Parvez, Md Masud; Basit, Abdul; Jariwala, Parth B.; Gáborik, Zsuzsanna; Kis, Emese; Heyward, Scott; Redinbo, Matthew R.; Prasad, Bhagwat (August 2021). "Quantitative Investigation of Irinotecan Metabolism, Transport, and Gut Microbiome Activation". Drug Metabolism and Disposition. 49 (8): 683–693. doi:10.1124/dmd.121.000476. PMC 8407663. PMID 34074730.
- ^ a b Sousa, Tiago; Paterson, Ronnie; Moore, Vanessa; Carlsson, Anders; Abrahamsson, Bertil; Basit, Abdul W. (November 2008). "The gastrointestinal microbiota as a site for the biotransformation of drugs". International Journal of Pharmaceutics. 363 (1–2): 1–25. doi:10.1016/j.ijpharm.2008.07.009. PMID 18682282.
- ^ a b Koppel, Nitzan; Bisanz, Jordan E; Pandelia, Maria-Eirini; Turnbaugh, Peter J; Balskus, Emily P (15 May 2018). "Discovery and characterization of a prevalent human gut bacterial enzyme sufficient for the inactivation of a family of plant toxins". eLife. 7. doi:10.7554/eLife.33953. PMC 5953540. PMID 29761785.
- ^ Alexander, James L.; Wilson, Ian D.; Teare, Julian; Marchesi, Julian R.; Nicholson, Jeremy K.; Kinross, James M. (June 2017). "Gut microbiota modulation of chemotherapy efficacy and toxicity". Nature Reviews Gastroenterology & Hepatology. 14 (6): 356–365. doi:10.1038/nrgastro.2017.20. hdl:10044/1/77636. PMID 28270698.
- ^ Brandi, Giovanni; Dabard, Jean; Raibaud, Pierre; Di Battista, Monica; Bridonneau, Chantal; Pisi, Anna Maria; Morselli Labate, Antonio Maria; Pantaleo, Maria Abbondanza; De Vivo, Antonello; Biasco, Guido (15 February 2006). "Intestinal microflora and digestive toxicity of irinotecan in mice". Clinical Cancer Research. 12 (4): 1299–1307. doi:10.1158/1078-0432.CCR-05-0750. PMID 16489087.
- ^ Koh, Ara; De Vadder, Filipe; Kovatcheva-Datchary, Petia; Bäckhed, Fredrik (June 2016). "From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites". Cell. 165 (6): 1332–1345. doi:10.1016/j.cell.2016.05.041. PMID 27259147.
- ^ Konopelski, Piotr; Ufnal, Marcin (14 September 2018). "Indoles - Gut Bacteria Metabolites of Tryptophan with Pharmacotherapeutic Potential". Current Drug Metabolism. 19 (10): 883–890. doi:10.2174/1389200219666180427164731. PMID 29708069.
- ^ a b Collins, Stephanie L.; Stine, Jonathan G.; Bisanz, Jordan E.; Okafor, C. Denise; Patterson, Andrew D. (April 2023). "Bile acids and the gut microbiota: metabolic interactions and impacts on disease". Nature Reviews Microbiology. 21 (4): 236–247. doi:10.1038/s41579-022-00805-x. PMID 36253479.
- ^ Yang, Wenjing; Yu, Tianming; Huang, Xiangsheng; Bilotta, Anthony J.; Xu, Leiqi; Lu, Yao; Sun, Jiaren; Pan, Fan; Zhou, Jia; Zhang, Wenbo; Yao, Suxia; Maynard, Craig L.; Singh, Nagendra; Dann, Sara M.; Liu, Zhanju; Cong, Yingzi (8 September 2020). "Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity". Nature Communications. 11 (1): 4457. Bibcode:2020NatCo..11.4457Y. doi:10.1038/s41467-020-18262-6. PMC 7478978. PMID 32901017.
- ^ Murugesan, Selvasankar; Nirmalkar, Khemlal; Hoyo-Vadillo, Carlos; García-Espitia, Matilde; Ramírez-Sánchez, Daniela; García-Mena, Jaime (April 2018). "Gut microbiome production of short-chain fatty acids and obesity in children". European Journal of Clinical Microbiology & Infectious Diseases. 37 (4): 621–625. doi:10.1007/s10096-017-3143-0. PMID 29196878.
- ^ a b Thanissery, Rajani; Winston, Jenessa A.; Theriot, Casey M. (June 2017). "Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids". Anaerobe. 45: 86–100. doi:10.1016/j.anaerobe.2017.03.004. PMC 5466893. PMID 28279860.
- ^ a b c Jones, Brian V.; Begley, Máire; Hill, Colin; Gahan, Cormac G. M.; Marchesi, Julian R. (9 September 2008). "Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome". Proceedings of the National Academy of Sciences. 105 (36): 13580–13585. Bibcode:2008PNAS..10513580J. doi:10.1073/pnas.0804437105. PMC 2533232. PMID 18757757.
- ^ a b Cao H, Xu M, Dong W, Deng B, Wang S, Zhang Y, Wang S, Luo S, Wang W, Qi Y, Gao J, Cao X, Yan F, Wang B (June 2017). "Secondary bile acid-induced dysbiosis promotes intestinal carcinogenesis". International Journal of Cancer. 140 (11): 2545–2556. doi:10.1002/ijc.30643. PMID 28187526.
- ^ a b Bernstein H, Bernstein C (January 2023). "Bile acids as carcinogens in the colon and at other sites in the gastrointestinal system". Experimental Biology and Medicine. 248 (1): 79–89. doi:10.1177/15353702221131858. PMC 9989147. PMID 36408538.
- ^ Mayer, E. A; Knight, R; Mazmanian, S. K; Cryan, J. F; Tillisch, K (2014). "Gut Microbes and the Brain: Paradigm Shift in Neuroscience". Journal of Neuroscience. 34 (46): 15490–15496. doi:10.1523/JNEUROSCI.3299-14.2014. PMC 4228144. PMID 25392516.
- ^ a b c Dinan, Timothy G; Cryan, John F (2015). "The impact of gut microbiota on brain and behaviour". Current Opinion in Clinical Nutrition and Metabolic Care. 18 (6): 552–558. doi:10.1097/MCO.0000000000000221. PMID 26372511.
- ^ a b c Carman, Robert J.; Simon, Mary Alice; Fernández, Haydée; Miller, Margaret A.; Bartholomew, Mary J. (2004). "Ciprofloxacin at low levels disrupts colonization resistance of human fecal microflora growing in chemostats". Regulatory Toxicology and Pharmacology. 40 (3): 319–326. doi:10.1016/j.yrtph.2004.08.005. PMID 15546686.
- ^ Hvas, Christian Lodberg; Baunwall, Simon Mark Dahl; Erikstrup, Christian (July 2020). "Faecal microbiota transplantation: A life-saving therapy challenged by commercial claims for exclusivity". eClinicalMedicine. 24: 100436. doi:10.1016/j.eclinm.2020.100436. PMC 7334803. PMID 32642633.
- ^ Brandt, Lawrence J.; Borody, Thomas Julius; Campbell, Jordana (2011). "Endoscopic Fecal Microbiota Transplantation". Journal of Clinical Gastroenterology. 45 (8): 655–657. doi:10.1097/MCG.0b013e3182257d4f. PMID 21716124.
- ^ Knight, DJW; Girling, KJ (2003). "Gut flora in health and disease". The Lancet. 361 (9371): 512–519. doi:10.1016/S0140-6736(03)13438-1. PMID 12781578.
- ^ a b c Cho, I.; Yamanishi, S.; Cox, L.; Methé, B. A.; Zavadil, J.; Li, K.; Gao, Z.; Mahana, D.; Raju, K.; Teitler, I.; Li, H.; Alekseyenko, A. V.; Blaser, M. J. (2012). "Antibiotics in early life alter the murine colonic microbiome and adiposity". Nature. 488 (7413): 621–626. Bibcode:2012Natur.488..621C. doi:10.1038/nature11400. PMC 3553221. PMID 22914093.
- ^ a b c Schneiderhan, J; Master-Hunter, T; Locke, A (2016). "Targeting gut flora to treat and prevent disease". The Journal of Family Practice. 65 (1): 34–38. PMID 26845162.
- ^ Baker, Monya (2012). "Pregnancy alters resident gut microbes". Nature. doi:10.1038/nature.2012.11118.
- ^ a b Horta-Baas, Gabriel; Sandoval-Cabrera, Antonio; Romero-Figueroa, María del Socorro (2021-07-03). "Modification of Gut Microbiota in Inflammatory Arthritis: Highlights and Future Challenges". Current Rheumatology Reports. 23 (8): 67. doi:10.1007/s11926-021-01031-9. ISSN 1534-6307. PMID 34218340.
- ^ Hill, Colin; Guarner, Francisco; Reid, Gregor; Gibson, Glenn R; Merenstein, Daniel J; Pot, Bruno; Morelli, Lorenzo; Canani, Roberto Berni; Flint, Harry J; Salminen, Seppo; Calder, Philip C; Sanders, Mary Ellen (2014). "The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic". Nature Reviews Gastroenterology & Hepatology. 11 (8): 506–514. doi:10.1038/nrgastro.2014.66. hdl:2164/4189. PMID 24912386.
- ^ Rijkers, Ger T; De Vos, Willem M; Brummer, Robert-Jan; Morelli, Lorenzo; Corthier, Gerard; Marteau, Philippe (2011). "Health benefits and health claims of probiotics: Bridging science and marketing". British Journal of Nutrition. 106 (9): 1291–1296. doi:10.1017/S000711451100287X. PMID 21861940.
- ^ Hutkins, Robert W; Krumbeck, Janina A; Bindels, Laure B; Cani, Patrice D; Fahey, George; Goh, Yong Jun; Hamaker, Bruce; Martens, Eric C; Mills, David A; Rastal, Robert A; Vaughan, Elaine; Sanders, Mary Ellen (2016). "Prebiotics: Why definitions matter". Current Opinion in Biotechnology. 37: 1–7. doi:10.1016/j.copbio.2015.09.001. PMC 4744122. PMID 26431716.
- ^ Pandey, Kavita. R; Naik, Suresh. R; Vakil, Babu. V (2015). "Probiotics, prebiotics and synbiotics- a review". Journal of Food Science and Technology. 52 (12): 7577–7587. doi:10.1007/s13197-015-1921-1. PMC 4648921. PMID 26604335.
- ^ Broeckx, Géraldine; Vandenheuvel, Dieter; Claes, Ingmar J.J; Lebeer, Sarah; Kiekens, Filip (2016). "Drying techniques of probiotic bacteria as an important step towards the development of novel pharmabiotics" (PDF). International Journal of Pharmaceutics. 505 (1–2): 303–318. doi:10.1016/j.ijpharm.2016.04.002. hdl:10067/1328840151162165141. PMID 27050865.
- ^ Sleator, Roy D; Hill, Colin (2009). "Rational Design of Improved Pharmabiotics". Journal of Biomedicine and Biotechnology. 2009: 275287. doi:10.1155/2009/275287. PMC 2742647. PMID 19753318.
- ^ Patterson, Elaine; Cryan, John F; Fitzgerald, Gerald F; Ross, R. Paul; Dinan, Timothy G; Stanton, Catherine (2014). "Gut microbiota, the pharmabiotics they produce and host health". Proceedings of the Nutrition Society. 73 (4): 477–489. doi:10.1017/S0029665114001426. PMID 25196939.
- ^ "Non-prescription therapeutics for IBS: Where are we?". 30 January 2024.
- ^ Guandalini, Stefano; Magazzù, Giuseppe; Chiaro, Andrea; La Balestra, Valeria; Di Nardo, Giovanni; Gopalan, Sarath; Sibal, A; Romano, Claudio; Canani, Roberto Berni; Lionetti, Paolo; Setty, Mala (July 2010). "VSL#3 Improves Symptoms in Children With Irritable Bowel Syndrome: A Multicenter, Randomized, Placebo-Controlled, Double-Blind, Crossover Study". Journal of Pediatric Gastroenterology and Nutrition. 51 (1): 24–30. doi:10.1097/MPG.0b013e3181ca4d95. PMID 20453678.
- ^ Crucillà, Salvatore; Caldart, Federico; Michelon, Marco; Marasco, Giovanni; Costantino, Andrea (2024-08-14). "Functional Abdominal Bloating and Gut Microbiota: An Update". Microorganisms. 12 (8): 1669. doi:10.3390/microorganisms12081669. PMC 11357468. PMID 39203511.
- ^ Ford, Alexander C; Quigley, Eamonn M M; Lacy, Brian E; Lembo, Anthony J; Saito, Yuri A; Schiller, Lawrence R; Soffer, Edy E; Spiegel, Brennan M R; Moayyedi, Paul (2014). "Efficacy of Prebiotics, Probiotics and Synbiotics in Irritable Bowel Syndrome and Chronic Idiopathic Constipation: Systematic Review and Meta-analysis". The American Journal of Gastroenterology. 109 (10): 1547–1561, quiz 1546, 1562. doi:10.1038/ajg.2014.202. PMID 25070051.
- ^ Dupont, Andrew; Richards; Jelinek, Katherine A; Krill, Joseph; Rahimi, Erik; Ghouri, Yezaz (2014). "Systematic review of randomized controlled trials of probiotics, prebiotics, and synbiotics in inflammatory bowel disease". Clinical and Experimental Gastroenterology. 7: 473–487. doi:10.2147/CEG.S27530. PMC 4266241. PMID 25525379.
- ^ Yu, Cheng Gong; Huang, Qin (2013). "Recent progress on the role of gut microbiota in the pathogenesis of inflammatory bowel disease". Journal of Digestive Diseases. 14 (10): 513–517. doi:10.1111/1751-2980.12087. PMID 23848393.
- ^ Levitt, Michael D.; Duane, William C. (4 May 1972). "Floating Stools — Flatus versus Fat". New England Journal of Medicine. 286 (18): 973–975. doi:10.1056/NEJM197205042861804. PMID 5015442.
- ^ Aalam, Syed Mohammed Musheer; Crasta, Daphne Norma; Roy, Pooja; Miller, A. Lee; Gamb, Scott I.; Johnson, Stephen; Till, Lisa M.; Chen, Jun; Kashyap, Purna; Kannan, Nagarajan (27 October 2022). "Genesis of fecal floatation is causally linked to gut microbial colonization in mice". Scientific Reports. 12 (1): 18109. Bibcode:2022NatSR..1218109A. doi:10.1038/s41598-022-22626-x. PMC 9613883. PMID 36302811.
- ^ Maier, Lisa; Pruteanu, Mihaela; Kuhn, Michael; Zeller, Georg; Telzerow, Anja; Anderson, Exene Erin; Brochado, Ana Rita; Fernandez, Keith Conrad; Dose, Hitomi; Mori, Hirotada; Patil, Kiran Raosaheb; Bork, Peer; Typas, Athanasios (2018). "Extensive impact of non-antibiotic drugs on human gut bacteria". Nature. 555 (7698): 623–628. Bibcode:2018Natur.555..623M. doi:10.1038/nature25979. PMC 6108420. PMID 29555994.
- ^ Clauss, Matthieu; Gérard, Philippe; Mosca, Alexis; Leclerc, Marion (2021). "Interplay Between Exercise and Gut Microbiome in the Context of Human Health and Performance". Frontiers in Nutrition. 8: 637010. doi:10.3389/fnut.2021.637010. PMC 8222532. PMID 34179053.
- ^ a b Kamboj, Amrit K.; Cotter, Thomas G.; Oxentenko, Amy S. (April 2017). "Helicobacter pylori". Mayo Clinic Proceedings. 92 (4): 599–604. doi:10.1016/j.mayocp.2016.11.017. PMID 28209367.
- ^ a b c "Peptic ulcer disease" (PDF). The Johns Hopkins University School of Medicine. 2013. Retrieved 21 October 2020.
- ^ Burisch, Johan; Jess, Tine; Martinato, Matteo; Lakatos, Peter L (2013). "The burden of inflammatory bowel disease in Europe". Journal of Crohn's and Colitis. 7 (4): 322–337. doi:10.1016/j.crohns.2013.01.010. PMID 23395397.
- ^ Blandino, G; Inturri, R; Lazzara, F; Di Rosa, M; Malaguarnera, L (2016). "Impact of gut microbiota on diabetes mellitus". Diabetes & Metabolism. 42 (5): 303–315. doi:10.1016/j.diabet.2016.04.004. PMID 27179626.
- ^ a b c Boulangé, Claire L; Neves, Ana Luisa; Chilloux, Julien; Nicholson, Jeremy K; Dumas, Marc-Emmanuel (2016). "Impact of the gut microbiota on inflammation, obesity, and metabolic disease". Genome Medicine. 8 (1): 42. doi:10.1186/s13073-016-0303-2. PMC 4839080. PMID 27098727.
- ^ a b c Arrieta, Marie-Claire; Stiemsma, Leah T; Dimitriu, Pedro A; Thorson, Lisa; Russell, Shannon; Yurist-Doutsch, Sophie; Kuzeljevic, Boris; Gold, Matthew J; Britton, Heidi M; Lefebvre, Diana L; Subbarao, Padmaja; Mandhane, Piush; Becker, Allan; McNagny, Kelly M; Sears, Malcolm R; Kollmann, Tobias; Mohn, William W; Turvey, Stuart E; Brett Finlay, B (2015). "Early infancy microbial and metabolic alterations affect risk of childhood asthma". Science Translational Medicine. 7 (307): 307ra152. doi:10.1126/scitranslmed.aab2271. PMID 26424567.
- ^ a b Stiemsma, Leah T; Turvey, Stuart E (2017). "Asthma and the microbiome: Defining the critical window in early life". Allergy, Asthma & Clinical Immunology. 13: 3. doi:10.1186/s13223-016-0173-6. PMC 5217603. PMID 28077947.
- ^ Ipci, Kagan; Altıntoprak, Niyazi; Muluk, Nuray Bayar; Senturk, Mehmet; Cingi, Cemal (2016). "The possible mechanisms of the human microbiome in allergic diseases". European Archives of Oto-Rhino-Laryngology. 274 (2): 617–626. doi:10.1007/s00405-016-4058-6. PMID 27115907.
- ^ Mariño, Eliana; Richards, James L; McLeod, Keiran H; Stanley, Dragana; Yap, Yu Anne; Knight, Jacinta; McKenzie, Craig; Kranich, Jan; Oliveira, Ana Carolina; Rossello, Fernando J; Krishnamurthy, Balasubramanian; Nefzger, Christian M; Macia, Laurence; Thorburn, Alison; Baxter, Alan G; Morahan, Grant; Wong, Lee H; Polo, Jose M; Moore, Robert J; Lockett, Trevor J; Clarke, Julie M; Topping, David L; Harrison, Leonard C; Mackay, Charles R (May 2017). "Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes". Nature Immunology. 18 (5): 552–562. doi:10.1038/ni.3713. PMID 28346408.
- ^ Bettelli, Estelle; Carrier, Yijun; Gao, Wenda; Korn, Thomas; Strom, Terry B.; Oukka, Mohamed; Weiner, Howard L.; Kuchroo, Vijay K. (May 2006). "Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells". Nature. 441 (7090): 235–238. doi:10.1038/nature04753. PMID 16648838.
- ^ a b Säemann, Marcus D.; Böhmig, Georg A.; Österreicher, Christoph H.; Burtscher, Helmut; Parolini, Ornella; Diakos, Christos; Stöckl, Johannes; Hörl, Walter H.; Zlabinger, Gerhard J. (December 2000). "Anti-inflammatory effects of sodium butyrate on human monocytes: potent inhibition of IL-12 and up-regulation of IL-10 production". The FASEB Journal. 14 (15): 2380–2382. doi:10.1096/fj.00-0359fje. PMID 11024006.
- ^ Gao, Z; Yin, J; Zhang, J; Ward, R. E; Martin, R. J; Lefevre, M; Cefalu, W. T; Ye, J (2009). "Butyrate Improves Insulin Sensitivity and Increases Energy Expenditure in Mice". Diabetes. 58 (7): 1509–1517. doi:10.2337/db08-1637. PMC 2699871. PMID 19366864.
- ^ Mazidi, Mohsen; Rezaie, Peyman; Kengne, Andre Pascal; Mobarhan, Majid Ghayour; Ferns, Gordon A (2016). "Gut microbiome and metabolic syndrome". Diabetes & Metabolic Syndrome: Clinical Research & Reviews. 10 (2): S150–157. doi:10.1016/j.dsx.2016.01.024. PMID 26916014.
- ^ Chen, Xiao; d'Souza, Roshan; Hong, Seong-Tshool (2013). "The role of gut microbiota in the gut-brain axis: Current challenges and perspectives". Protein & Cell. 4 (6): 403–414. doi:10.1007/s13238-013-3017-x. PMC 4875553. PMID 23686721.
- ^ a b Moeller, Andrew H.; Li, Yingying; Mpoudi Ngole, Eitel; Ahuka-Mundeke, Steve; Lonsdorf, Elizabeth V.; Pusey, Anne E.; Peeters, Martine; Hahn, Beatrice H.; Ochman, Howard (18 November 2014). "Rapid changes in the gut microbiome during human evolution". Proceedings of the National Academy of Sciences. 111 (46): 16431–16435. Bibcode:2014PNAS..11116431M. doi:10.1073/pnas.1419136111. PMC 4246287. PMID 25368157.
- ^ Brune, A. (2014). "Symbiotic digestion of lignocellulose in termite guts". Nature Reviews Microbiology. 12 (3): 168–180. doi:10.1038/nrmicro3182. PMID 24487819.
- ^ Rosengaus, Rebeca B.; Zecher, Courtney N.; Schultheis, Kelley F.; Brucker, Robert M.; Bordenstein, Seth R. (July 2011). "Disruption of the Termite Gut Microbiota and Its Prolonged Consequences for Fitness". Applied and Environmental Microbiology. 77 (13): 4303–4312. Bibcode:2011ApEnM..77.4303R. doi:10.1128/AEM.01886-10. PMC 3127728. PMID 21571887.
- ^ a b Ashbrook, Aaron R; Schwarz, Melbert; Schal, Coby; Mikaelyan, Aram (14 October 2024). "Lethal disruption of the bacterial gut community in Eastern subterranean termite caused by boric acid". Journal of Economic Entomology. doi:10.1093/jee/toae221. PMID 39401329.
- ^ a b Dietrich, C.; Köhler, T.; Brune, A. (2014). "The cockroach origin of the termite gut microbiota: patterns in bacterial community structure reflect major evolutionary events". Applied and Environmental Microbiology. 80 (7): 2261–2269. Bibcode:2014ApEnM..80.2261D. doi:10.1128/AEM.04206-13. PMC 3993134. PMID 24487532.
- ^ a b Mikaelyan, A.; Dietrich, C.; Köhler, T.; Poulsen, M.; Sillam-Dussès, D.; Brune, A. (2015). "Diet is the primary determinant of bacterial community structure in the guts of higher termites". Molecular Ecology. 24 (20): 5824–5895. Bibcode:2015MolEc..24.5284M. doi:10.1111/mec.13376. PMID 26348261.
- ^ Mikaelyan, A.; Thompson, C.; Hofer, M.; Brune, A. (2016). "The deterministic assembly of complex bacterial communities in germ-free cockroach guts". Applied and Environmental Microbiology. 82 (4): 1256–1263. doi:10.1128/AEM.03700-15. PMC 4751828. PMID 26655763.
Further reading
[edit]- Review articles
- De Preter, Vicky; Hamer, Henrike M; Windey, Karen; Verbeke, Kristin (2011). "The impact of pre- and/or probiotics on human colonic metabolism: Does it affect human health?". Molecular Nutrition & Food Research. 55 (1): 46–57. doi:10.1002/mnfr.201000451. PMID 21207512.
- Maranduba, Carlos Magno da Costa; De Castro, Sandra Bertelli Ribeiro; Souza, Gustavo Torres de; Rossato, Cristiano; Da Guia, Francisco Carlos; Valente, Maria Anete Santana; Rettore, João Vitor Paes; Maranduba, Claudinéia Pereira; Souza, Camila Maurmann de; Carmo, Antônio Márcio Resende do; MacEdo, Gilson Costa; Silva, Fernando de Sá (2015). "Intestinal Microbiota as Modulators of the Immune System and Neuroimmune System: Impact on the Host Health and Homeostasis". Journal of Immunology Research. 2015: 931574. doi:10.1155/2015/931574. PMC 4352473. PMID 25759850.
- Prakash, Satya; Rodes, Laetitia; Coussa-Charley, Michael; Tomaro-Duchesneau, Catherine; Tomaro-Duchesneau, Catherine; Coussa-Charley; Rodes (2011). "Gut microbiota: Next frontier in understanding human health and development of biotherapeutics". Biologics: Targets and Therapy. 5: 71–86. doi:10.2147/BTT.S19099. PMC 3156250. PMID 21847343.
- Wu, G. D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S. A.; Bewtra, M.; Knights, D.; Walters, W. A.; Knight, R.; Sinha, R.; Gilroy, E.; Gupta, K.; Baldassano, R.; Nessel, L.; Li, H.; Bushman, F. D.; Lewis, J.D. (2011). "Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes". Science. 334 (6052): 105–108. Bibcode:2011Sci...334..105W. doi:10.1126/science.1208344. PMC 3368382. PMID 21885731.