G1/S transition


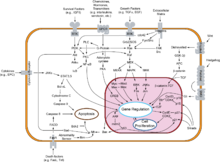
The G1/S transition is a stage in the cell cycle at the boundary between the G1 phase, in which the cell grows, and the S phase, during which DNA is replicated.[1] It is governed by cell cycle checkpoints to ensure cell cycle integrity and the subsequent S phase can pause in response to improperly or partially replicated DNA.[2] During this transition the cell makes decisions to become quiescent (enter G0), differentiate, make DNA repairs, or proliferate based on environmental cues and molecular signaling inputs.[3] The G1/S transition occurs late in G1 and the absence or improper application of this highly regulated checkpoint can lead to cellular transformation and disease states such as cancer.[4][1]
During this transition, G1 cyclin D-Cdk4/6 dimer phosphorylates retinoblastoma releasing transcription factor E2F, which then drives the transition from G1 to S phase. The G1/S transition is highly regulated by transcription factor p53 in order to halt the cell cycle when DNA is damaged.[5]
It is a "point of no return" beyond which the cell is committed to dividing; in yeast this is called the Start point, and in multicellular eukaryotes it is termed the restriction point (R-Point).[2][6] If a cell passes through the G1/S transition the cell will continue through the cell cycle regardless of incoming mitogenic factors due to the positive feed-back loop of G1-S transcription.[2] Positive feed-back loops include G1 cyclins and accumulation of E2F in multicellular eukaryotes, and the accumulation of SBF in yeast cells.[2]
Cell cycle overview
[edit]The cell cycle is a process in which an ordered set of events leads to the growth and division into two daughter cells. The cell cycle is a cycle rather than a linear process because the two daughter cells produced repeat the cycle. This process contains two main phases, interphase, in which the cell grows and synthesizes a copy of its DNA, and the mitotic (M) phase, during which the cell separates its DNA and divides into two new daughter cells.[7] Interphase is further broken down into the G1 (GAP 1) phase, S (Synthesis) phase, G2 (GAP 2) phase and the mitotic (M) phase which in turn is broken down into mitosis and cytokinesis. Following cytokinesis, during G1 phase the cells monitor environment for the potential growth factors, grow larger and once achieve the threshold size (rRNA and overall protein content characteristic for a given cell type) they start progression through S phase.[8] During S phase, the cell also duplicates the centrosome, or microtubule-organizing center, which is critical for DNA separation in the M phase. After complete synthesis of its DNA, the cell enters the G2 phase where it continues to grow in preparation for mitosis. Following interphase, the cell transitions into mitosis, containing four sub stages: prophase, anaphase, metaphase, and telophase. In mitosis, DNA condenses into chromosomes, which are lined up and separated by the mitotic spindle.[9] After duplicate DNA is separated on opposite ends of the cell, the cytoplasm of the cell is split in two during cytokinesis resulting in two daughter cells.[7]
The yeast cell cycle goes through similar stages however there is the additional factor of mating to consider. A haploid cell arrests in G1 if it has not passed Start and is exposed to enough mating pheromone, but will progress into S-phase if both of those conditions are not met.[10]
Cell cycle regulation in mammalian cells
[edit]As with most processes in the body, the cell cycle is highly regulated to prevent the synthesis of mutated cells and uncontrolled cell division that leads to tumor formation.[11] The cell cycle control system is biochemically based so that the proteins of the mitosis promoting factor (MPF) control the transition from one phase to the next based on a series of checkpoints. MPF is a protein dimer made up of cyclin and cyclin-dependent kinase (Cdk), a serine and threonine kinase, which come together at different points in the cycle to control cell progression through the cycle. When cyclin binds to Cdk, Cdk becomes activated and phosphorylates serine and threonine on other proteins causing the activation and degradation of other proteins allowing the cell to transition through the cell cycle.[7]
G1/S transition
[edit]In mid to late G1 phase, cyclin D bound to Cdk4/6, activates the expression of the S phase cyclin-Cdk components; however, the cell does not want S phase cyclins to become active in G1.[7] Therefore, an inhibitor, protein Slc-1, is present that interacts with the dimer so that the S phase cyclin-Cdk dimer remains inactive until the cell is ready to move into S phase.[7] After the cell has grown and is ready to synthesize DNA, G1 cyclin-Cdks phosphorylate the S phase cyclin inhibitor signaling ubiquitination, resulting in the addition of groups to the inhibitor. Ubiquitination of the inhibitor signals the SCF/proteasome to degrade the inhibitor releasing and allowing the S phase cyclin-Cdk to become activated and the cell moves into S phase. Once in S phase, cyclin-Cdks phosphorylate several factors on the replication complex promoting DNA replication by causing inhibitory proteins to fall off of replication complexes or through activation of components on the replication complex to induce DNA replication initiation.[12]
Retinoblastoma protein (pRB) and the G1/S transition
[edit]
Another dimer present during mid G1 is composed of retinoblastoma protein (pRB) and transcription factor E2F. When pRb is bound to E2F, E2F is inactive. As cyclin D is synthesized and activates Cdk4/6, the cyclin-Cdk targets Rb protein for phosphorylation. Upon phosphorylation, pRb changes conformation so that E2F is released and activated, binding to upstream regions of genes, initiating expression. Specifically, E2F drives the expression of other cyclins, including cyclin E and A, and genes necessary for DNA replication. Cyclin E either phosphorylates more pRb, thereby completing inactivation, to further activate E2F and promote the expression of more Cyclin E, or it has the ability to increase expression of itself. These interactions create a positive feedback loop. Cyclin E also interacts with Cdk2 driving the cell cycle to progress from G1 to S phase.[13] E2F also targets Skp2, an F-box protein that targets the CDK inhibitor p27 for degradation, creating a third positive feedback loop. These positive feedback loops are the key to creating the all-or-nothing, switch-like transition between G1 and S-phase, and their components cyclin E1, cyclin E2, Skp2, and E2F1 are all transcribed early on at the G1/S transition.[14]
The role of retinoblastoma in tumor formation
[edit]Retinoblastoma (Rb) is a cancer of the eye due to a mutant pRb protein.[7] When pRb is mutated it becomes nonfunctional and is not able to inhibit the expression of transcription factor E2F. Therefore, E2F is always active and driving the cell cycle to progress from G1 to S phase. As a result, cell growth and division is unregulated causing tumor formation in the eye.[11]
Cell cycle checkpoints
[edit]To ensure proper cell division, the cell cycle utilizes numerous checkpoints to monitor cell progression and halt the cycle when processes go awry. These checkpoints include four DNA damage checkpoints, one unreplicated DNA checkpoint at the end of G2, one spindle assembly checkpoint in mitosis, and a chromosome segregation checkpoint during mitosis.[11]
p53 as a regulator
[edit]

Between G1 and S phase, three DNA damage checkpoints occur to ensure proper growth and synthesis of DNA prior to cell division. Damaged DNA during G1, before entry into S phase, and during S phase result in the expression of ATM/R protein. ATM/R protein then stabilizes and activates transcription factor p53 so that it can bind to upstream regions of genes, inducing the expression of proteins including p21CIP. p21CIP binds to and inhibits any cyclin-cdk present in the cell cycle, halting the cycle until DNA damage can be corrected.[15]
Additional processes at DNA damage checkpoints
[edit]Of the four DNA damage checkpoints, two have an additional process for monitoring DNA damage other than activating p53. Before entry into S phase and during S phase, ATM/R also activates Chk1/2 that inhibits Cdc25A, a protein responsible for activating cyclin-Cdk dimers. Without cyclin dimer activation, the cell cannot transition through the cycle. These two checkpoints have additional processes for regulation because replicating damaged DNA in S phase can be deleterious to the cell and more importantly, the organism.[7]
Cell cycle regulation in budding yeast
[edit]Cell cycle regulation is just as important in yeast cells which respond to nutrient and mating pheromone levels in their environment in order to grow, divide and reproduce appropriately.
The Start point is the moment in the yeast cell cycle that determines the all-or-nothing commitment to undergo DNA replication. Input signals promote cyclin synthesis which drives cyclin-dependent kinase (CDK) activity past a threshold that triggers a self-sustaining positive feedback loop that pushes the cell into S-phase.[14] This is a sharp switch due to the genes involved in the positive feedback loop being transcribed before other genes transcribed under the same transcriptional factor.
G1/S transition
[edit]Cln3 binds and activates CDK1. The Cln3-CDK1 complex then inactivates the transcriptional inhibitor Whi5 by phosphorylation which causes it to release from the transcriptional factor SBF. The size of a yeast cell and its time spent in G1 are roughly proportional. This is partly governed by inhibitor dilution associated with Whi5. Once no longer inhibited, SBF goes on to weakly promote transcription of downstream G1 cyclins CLN1 and CLN2. Cln1 and Cln2 form a positive feedback loop by further inactivating Whi5 and concurrently activating SBF as well as MBF, driving the expression of over 200 genes, including S-phase cyclins that initiate DNA replication. An additional positive feedback loop is created by Swi4, a component of SBF that is itself a target of SBF. Together, this sudden activation of the G1 cyclin positive feedback loop defines the Start point that is key to making the G1/S transition distinct and abrupt. Once the CDK activity threshold is passed and feedback is activated, fluctuations in the upstream input signals no longer have an influence on the fate of the cell cycle.[14]
See also
[edit]References
[edit]- ^ a b Bartek J, Lukas J (February 2001). "Pathways governing G1/S transition and their response to DNA damage". FEBS Letters. 490 (3): 117–22. Bibcode:2001FEBSL.490..117B. doi:10.1016/S0014-5793(01)02114-7. PMID 11223026. S2CID 16090531.
- ^ a b c d Bertoli C, Skotheim JM, de Bruin RA (August 2013). "Control of cell cycle transcription during G1 and S phases". Nature Reviews Molecular Cell Biology. 14 (8): 518–28. doi:10.1038/nrm3629. PMC 4569015. PMID 23877564.
- ^ Massagué J (November 2004). "G1 cell-cycle control and cancer". Nature. 432 (7015): 298–306. Bibcode:2004Natur.432..298M. doi:10.1038/nature03094. PMID 15549091. S2CID 4428026.
- ^ Bartek J, Lukas J (February 2001). "Pathways governing G1/S transition and their response to DNA damage". FEBS Letters. 490 (3): 117–22. Bibcode:2001FEBSL.490..117B. doi:10.1016/S0014-5793(01)02114-7. PMID 11223026. S2CID 16090531.
- ^ Lodish H, Berk A, Kaiser C, Krieger M (2012). Molecular Cell Biology (7th ed.). Freeman, W. H. & Company. ISBN 978-1-4641-0981-2.
- ^ Tenga MJ, Lazar IM (January 2013). "Proteomic snapshot of breast cancer cell cycle: G1/S transition point". Proteomics. 13 (1): 48–60. doi:10.1002/pmic.201200188. PMC 4123745. PMID 23152136.
- ^ a b c d e f g Lodish H, Berk A, Kaiser C, Krieger M (2012). Molecular Cell Biology (7th 13 ed.). Freeman, W. H. & Company. ISBN 978-1-4641-0981-2.
- ^ Darzynkiewicz, Z; Sharpless, T; Staiano-Coico, L; Melamed, MR (1980). "Subcompartments of the G1 phase of cell cycle detected by flow cytometry". Proceedings of the National Academy of Sciences of the United States of America. 77 (11): 6696–9. Bibcode:1980PNAS...77.6696D. doi:10.1073/pnas.77.11.6696. PMC 350355. PMID 6161370.
- ^ "Phases of the Cell Cycle". KhanAcademy.
- ^ Yeager, Randi; Bushkin, G. Guy; Singer, Emily; Fu, Rui; Cooperman, Benjamin; McMurray, Michael (2021-08-17). "Post-Transcriptional Control of Mating-Type Gene Expression during Gametogenesis in Saccharomyces cerevisiae". Biomolecules. 11 (8): 1223. doi:10.3390/biom11081223. ISSN 2218-273X. PMC 8394074. PMID 34439889.
- ^ a b c Alao JP (April 2007). "The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention". Molecular Cancer. 6: 24. doi:10.1186/1476-4598-6-24. PMC 1851974. PMID 17407548.
- ^ Poli A (2015). New DAG-dependent mechanisms modulate cell cycle progression [Dissertation]. Scienze Biomediche (Doctoral Thesis). doi:10.6092/unibo/amsdottorato/6739.
- ^ Fadila, Guessous; Jinho, Heo; Vaddadi, Naga; Abbas, Tarek (2015). "Abstract 3786: Novel regulation of cyclin D1 stability and the DNA damage response". Cancer Research. 75 (15 Supplement): 3786. doi:10.1158/1538-7445.AM2015-3786.
- ^ a b c Johnson, Amy; Skotheim, Jan M (2013-12-01). "Start and the restriction point". Current Opinion in Cell Biology. Cell cycle, differentiation and disease. 25 (6): 717–723. doi:10.1016/j.ceb.2013.07.010. ISSN 0955-0674. PMC 3836907. PMID 23916770.
- ^ Wang X, Simpson ER, Brown KA (December 2015). "p53: Protection against Tumor Growth beyond Effects on Cell Cycle and Apoptosis". Cancer Research. 75 (23): 5001–7. doi:10.1158/0008-5472.CAN-15-0563. PMID 26573797.
