Fatty acid synthase
| Fatty acid synthase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 2.3.1.85 | ||||||||
| CAS no. | 9045-77-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| fatty acid synthase | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | fatty acid synthases | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | GeneCards: [1]; OMA:- orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Fatty acid synthase (FAS)[1] is an enzyme that in humans is encoded by the FASN gene.[2][3][4][5]
Fatty acid synthase is a multi-enzyme protein that catalyzes fatty acid synthesis. It is not a single enzyme but a whole enzymatic system composed of two identical 272 kDa multifunctional polypeptides, in which substrates are handed from one functional domain to the next.[1][6][7][8][9]
Its main function is to catalyze the synthesis of palmitate (C16:0, a long-chain saturated fatty acid) from acetyl-CoA and malonyl-CoA, in the presence of NADPH.[5]
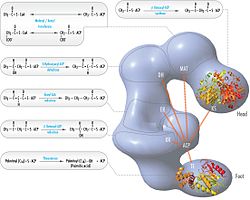
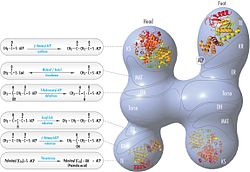
The fatty acids are synthesized by a series of decarboxylative Claisen condensation reactions from acetyl-CoA and malonyl-CoA. Following each round of elongation the beta keto group is reduced to the fully saturated carbon chain by the sequential action of a ketoreductase (KR), dehydratase (DH), and enoyl reductase (ER). The growing fatty acid chain is carried between these active sites while attached covalently to the phosphopantetheine prosthetic group of an acyl carrier protein (ACP), and is released by the action of a thioesterase (TE) upon reaching a carbon chain length of 16 (palmitic acid).[1]
Classes
[edit]There are two principal classes of fatty acid synthases.
- Type I systems utilise a single large, multifunctional polypeptide and are common to both animals and fungi (although the structural arrangement of fungal and animal syntheses differ). A Type I fatty acid synthase system is also found in the CMN group of bacteria (corynebacteria, mycobacteria, and nocardia). In these bacteria, the FAS I system produces palmitic acid, and cooperates with the FAS II system to produce a greater diversity of lipid products.[10]
- Type II is found in archaea, bacteria and plant plastids, and is characterized by the use of discrete, monofunctional enzymes for fatty acid synthesis. Inhibitors of this pathway (FASII) are being investigated as possible antibiotics.[11]
The mechanism of FAS I and FAS II elongation and reduction is the same, as the domains of the FAS II enzymes are largely homologous to their domain counterparts in FAS I multienzyme polypeptides. However, the differences in the organization of the enzymes - integrated in FAS I, discrete in FAS II - gives rise to many important biochemical differences.[12]
The evolutionary history of fatty acid synthases are very much intertwined with that of polyketide synthases (PKS). Polyketide synthases use a similar mechanism and homologous domains to produce secondary metabolite lipids. Furthermore, polyketide synthases also exhibit a Type I and Type II organization. FAS I in animals is thought to have arisen through modification of PKS I in fungi, whereas FAS I in fungi and the CMN group of bacteria seem to have arisen separately through the fusion of FAS II genes.[10]
Structure
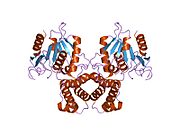
[edit]Mammalian FAS consists of a homodimer of two identical protein subunits, in which three catalytic domains in the N-terminal section (-ketoacyl synthase (KS), malonyl/acetyltransferase (MAT), and dehydrase (DH)), are separated by a core region (known as the interdomain) of 600 residues from four C-terminal domains (enoyl reductase (ER), -ketoacyl reductase (KR), acyl carrier protein (ACP) and thioesterase (TE)).[13][14] The interdomain region allows the two monomeric domains to form a dimer.[13]
The conventional model for organization of FAS (see the 'head-to-tail' model on the right) is largely based on the observations that the bifunctional reagent 1,3-dibromopropanone (DBP) is able to crosslink the active site cysteine thiol of the KS domain in one FAS monomer with the phosphopantetheine prosthetic group of the ACP domain in the other monomer.[15][16] Complementation analysis of FAS dimers carrying different mutations on each monomer has established that the KS and MAT domains can cooperate with the ACP of either monomer.[17][18] and a reinvestigation of the DBP crosslinking experiments revealed that the KS active site Cys161 thiol could be crosslinked to the ACP 4'-phosphopantetheine thiol of either monomer.[19] In addition, it has been recently reported that a heterodimeric FAS containing only one competent monomer is capable of palmitate synthesis.[20]
The above observations seemed incompatible with the classical 'head-to-tail' model for FAS organization, and an alternative model has been proposed, predicting that the KS and MAT domains of both monomers lie closer to the center of the FAS dimer, where they can access the ACP of either subunit (see figure on the top right).[21]
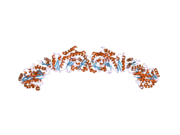
A low resolution X-ray crystallography structure of both pig (homodimer)[22] and yeast FAS (heterododecamer)[23] along with a ~6 Å resolution electron cryo-microscopy (cryo-EM) yeast FAS structure [24] have been solved.
Substrate shuttling mechanism
[edit]The solved structures of yeast FAS and mammalian FAS show two distinct organizations of highly conserved catalytic domains/enzymes in this multi-enzyme cellular machine. Yeast FAS has a highly efficient rigid barrel-like structure with 6 reaction chambers which synthesize fatty acids independently, while the mammalian FAS has an open flexible structure with only two reaction chambers. However, in both cases the conserved ACP acts as the mobile domain responsible for shuttling the intermediate fatty acid substrates to various catalytic sites. A first direct structural insight into this substrate shuttling mechanism was obtained by cryo-EM analysis, where ACP is observed bound to the various catalytic domains in the barrel-shaped yeast fatty acid synthase.[24] The cryo-EM results suggest that the binding of ACP to various sites is asymmetric and stochastic, as also indicated by computer-simulation studies[25]
 |
 |
Regulation
[edit]Metabolism and homeostasis of fatty acid synthase is transcriptionally regulated by Upstream Stimulatory Factors (USF1 and USF2) and sterol regulatory element binding protein-1c (SREBP-1c) in response to feeding/insulin in living animals.[26][27]
Although liver X receptors (LXRs) modulate the expression of sterol regulatory element binding protein-1c (SREBP-1c) in feeding, regulation of FAS by SREBP-1c is USF-dependent.[27][28][29][30]
Acylphloroglucinols isolated from the fern Dryopteris crassirhizoma show a fatty acid synthase inhibitory activity.[31]
Clinical significance
[edit]The FASN gene has been investigated as a possible oncogene.[32] FAS is upregulated in breast and gastric cancers, as well as being an indicator of poor prognosis, and so may be worthwhile as a chemotherapeutic target.[33][34][35] FAS inhibitors are therefore an active area of drug discovery research.[36][37][38][39][40]
FAS may also be involved in the production of an endogenous ligand for the nuclear receptor PPARalpha, the target of the fibrate drugs for hyperlipidemia,[41] and is being investigated as a possible drug target for treating the metabolic syndrome.[42] Orlistat which is a gastrointestinal lipase inhibitor also inhibits FAS and has a potential as a medicine for cancer.[43][44]
In some cancer cell lines, this protein has been found to be fused with estrogen receptor alpha (ER-alpha), in which the N-terminus of FAS is fused in-frame with the C-terminus of ER-alpha.[5]
An association with uterine leiomyomata has been reported.[45]
See also
[edit]- Discovery and development of gastrointestinal lipase inhibitors
- Fatty acid synthesis
- Fatty acid metabolism
- Fatty acid degradation
- Enoyl-acyl carrier protein reductase
- List of fatty acid metabolism disorders
References
[edit]- ^ a b c Paiva P, Medina FE, Viegas M, Ferreira P, Neves RP, Sousa JP, Ramos MJ, Fernandes PA (2021-08-11). "Animal Fatty Acid Synthase: A Chemical Nanofactory". Chemical Reviews. 121 (15): 9502–9553. doi:10.1021/acs.chemrev.1c00147. ISSN 0009-2665. PMID 34156235. S2CID 235595027.
- ^ Jayakumar A, Chirala SS, Chinault AC, Baldini A, Abu-Elheiga L, Wakil SJ (February 1995). "Isolation and chromosomal mapping of genomic clones encoding the human fatty acid synthase gene". Genomics. 23 (2): 420–424. doi:10.1006/geno.1994.1518. PMID 7835891.
- ^ Jayakumar A, Tai MH, Huang WY, al-Feel W, Hsu M, Abu-Elheiga L, Chirala SS, Wakil SJ (Oct 1995). "Human fatty acid synthase: properties and molecular cloning". Proceedings of the National Academy of Sciences of the United States of America. 92 (19): 8695–8699. Bibcode:1995PNAS...92.8695J. doi:10.1073/pnas.92.19.8695. PMC 41033. PMID 7567999.
- ^ Persson B, Kallberg Y, Bray JE, Bruford E, Dellaporta SL, Favia AD, Duarte RG, Jörnvall H, Kavanagh KL, Kedishvili N, Kisiela M, Maser E, Mindnich R, Orchard S, Penning TM, Thornton JM, Adamski J, Oppermann U (Feb 2009). "The SDR (short-chain dehydrogenase/reductase and related enzymes) nomenclature initiative". Chemico-Biological Interactions. 178 (1–3): 94–98. Bibcode:2009CBI...178...94P. doi:10.1016/j.cbi.2008.10.040. PMC 2896744. PMID 19027726.
- ^ a b c "Entrez Gene: FASN fatty acid synthase".
- ^ Alberts AW, Strauss AW, Hennessy S, Vagelos PR (October 1975). "Regulation of synthesis of hepatic fatty acid synthetase: binding of fatty acid synthetase antibodies to polysomes". Proceedings of the National Academy of Sciences of the United States of America. 72 (10): 3956–3960. Bibcode:1975PNAS...72.3956A. doi:10.1073/pnas.72.10.3956. PMC 433116. PMID 1060077.
- ^ Stoops JK, Arslanian MJ, Oh YH, Aune KC, Vanaman TC, Wakil SJ (May 1975). "Presence of two polypeptide chains comprising fatty acid synthetase". Proceedings of the National Academy of Sciences of the United States of America. 72 (5): 1940–1944. Bibcode:1975PNAS...72.1940S. doi:10.1073/pnas.72.5.1940. PMC 432664. PMID 1098047.
- ^ Smith S, Agradi E, Libertini L, Dileepan KN (April 1976). "Specific release of the thioesterase component of the fatty acid synthetase multienzyme complex by limited trypsinization". Proceedings of the National Academy of Sciences of the United States of America. 73 (4): 1184–1188. Bibcode:1976PNAS...73.1184S. doi:10.1073/pnas.73.4.1184. PMC 430225. PMID 1063400.
- ^ Smith S, Witkowski A, Joshi AK (July 2003). "Structural and functional organization of the animal fatty acid synthase". Progress in Lipid Research. 42 (4): 289–317. doi:10.1016/S0163-7827(02)00067-X. PMID 12689621.
- ^ a b Jenke-Kodama H, Sandmann A, Müller R, Dittmann E (October 2005). "Evolutionary implications of bacterial polyketide synthases". Molecular Biology and Evolution. 22 (10): 2027–2039. doi:10.1093/molbev/msi193. PMID 15958783.
- ^ Fulmer T (March 2009). "Not so FAS". Science-Business EXchange. 2 (11): 430. doi:10.1038/scibx.2009.430.
- ^ Stevens L, Price NC (1999). Fundamentals of enzymology: the cell and molecular biology of catalytic proteins. Oxford [Oxfordshire]: Oxford University Press. ISBN 978-0-19-850229-6.
- ^ a b Chirala SS, Jayakumar A, Gu ZW, Wakil SJ (March 2001). "Human fatty acid synthase: role of interdomain in the formation of catalytically active synthase dimer". Proceedings of the National Academy of Sciences of the United States of America. 98 (6): 3104–3108. Bibcode:2001PNAS...98.3104C. doi:10.1073/pnas.051635998. PMC 30614. PMID 11248039.
- ^ Smith S (December 1994). "The animal fatty acid synthase: one gene, one polypeptide, seven enzymes". FASEB Journal. 8 (15): 1248–1259. doi:10.1096/fasebj.8.15.8001737. PMID 8001737. S2CID 22853095.
- ^ Stoops JK, Wakil SJ (May 1981). "Animal fatty acid synthetase. A novel arrangement of the beta-ketoacyl synthetase sites comprising domains of the two subunits". Journal of Biological Chemistry. 256 (10): 5128–5133. doi:10.1016/S0021-9258(19)69376-2. PMID 6112225.
- ^ Stoops JK, Wakil SJ (March 1982). "Animal fatty acid synthetase. Identification of the residues comprising the novel arrangement of the beta-ketoacyl synthetase site and their role in its cold inactivation". Journal of Biological Chemistry. 257 (6): 3230–3235. doi:10.1016/S0021-9258(19)81100-6. PMID 7061475.
- ^ Joshi AK, Rangan VS, Smith S (February 1998). "Differential affinity labeling of the two subunits of the homodimeric animal fatty acid synthase allows isolation of heterodimers consisting of subunits that have been independently modified". Journal of Biological Chemistry. 273 (9): 4937–4943. doi:10.1074/jbc.273.9.4937. PMID 9478938.
- ^ Rangan VS, Joshi AK, Smith S (September 2001). "Mapping the functional topology of the animal fatty acid synthase by mutant complementation in vitro". Biochemistry. 40 (36): 10792–18799. doi:10.1021/bi015535z. PMID 11535054.
- ^ Witkowski A, Joshi AK, Rangan VS, Falick AM, Witkowska HE, Smith S (April 1999). "Dibromopropanone cross-linking of the phosphopantetheine and active-site cysteine thiols of the animal fatty acid synthase can occur both inter- and intrasubunit. Reevaluation of the side-by-side, antiparallel subunit model". Journal of Biological Chemistry. 274 (17): 11557–11563. doi:10.1074/jbc.274.17.11557. PMID 10206962.
- ^ Joshi AK, Rangan VS, Witkowski A, Smith S (February 2003). "Engineering of an active animal fatty acid synthase dimer with only one competent subunit". Chemistry and Biology. 10 (2): 169–173. doi:10.1016/S1074-5521(03)00023-1. PMID 12618189.
- ^ Asturias FJ, Chadick JZ, Cheung IK, Stark H, Witkowski A, Joshi AK, Smith S (March 2005). "Structure and molecular organization of mammalian fatty acid synthase". Nature Structural and Molecular Biology. 12 (3): 225–232. doi:10.1038/nsmb899. PMID 15711565. S2CID 6132878.
- ^ Maier T, Leibundgut M, Ban N (September 2008). "The crystal structure of a mammalian fatty acid synthase". Science. 321 (5894): 1315–1322. Bibcode:2008Sci...321.1315M. doi:10.1126/science.1161269. PMID 18772430. S2CID 3168991.
- ^ Lomakin IB, Xiong Y, Steitz TA (April 2007). "The crystal structure of yeast fatty acid synthase, a cellular machine with eight active sites working together". Cell. 129 (2): 319–332. doi:10.1016/j.cell.2007.03.013. PMID 17448991. S2CID 8209424.
- ^ a b Gipson P, Mills DJ, Wouts R, Grininger M, Vonck J, Kühlbrandt W (May 2010). "Direct structural insight into the substrate-shuttling mechanism of yeast fatty acid synthase by electron cryomicroscopy". Proceedings of the National Academy of Sciences of the United States of America. 107 (20): 9164–9169. Bibcode:2010PNAS..107.9164G. doi:10.1073/pnas.0913547107. PMC 2889056. PMID 20231485.
- ^ Anselmi C, Grininger M, Gipson P, Faraldo-Gómez JD (September 2010). "Mechanism of substrate shuttling by the acyl-carrier protein within the fatty acid mega-synthase". Journal of the American Chemical Society. 132 (35): 12357–12364. doi:10.1021/ja103354w. PMID 20704262.
- ^ Paulauskis JD, Sul HS (January 1989). "Hormonal regulation of mouse fatty acid synthase gene transcription in liver". Journal of Biological Chemistry. 264 (1): 574–577. doi:10.1016/S0021-9258(17)31298-X. PMID 2535847.
- ^ a b Latasa MJ, Griffin MJ, Moon YS, Kang C, Sul HS (August 2003). "Occupancy and function of the -150 sterol regulatory element and -65 E-box in nutritional regulation of the fatty acid synthase gene in living animals". Molecular and Cellular Biology. 23 (16): 5896–5907. doi:10.1128/MCB.23.16.5896-5907.2003. PMC 166350. PMID 12897158.
- ^ Griffin MJ, Wong RH, Pandya N, Sul HS (February 2007). "Direct interaction between USF and SREBP-1c mediates synergistic activation of the fatty-acid synthase promoter". Journal of Biological Chemistry. 282 (8): 5453–5467. doi:10.1074/jbc.M610566200. PMID 17197698.
- ^ Yoshikawa T, Shimano H, Amemiya-Kudo M, Yahagi N, Hasty AH, Matsuzaka T, Okazaki H, Tamura Y, Iizuka Y, Ohashi K, Osuga J, Harada K, Gotoda T, Kimura S, Ishibashi S, Yamada N (May 2001). "Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter". Molecular and Cellular Biology. 21 (9): 2991–3000. doi:10.1128/MCB.21.9.2991-3000.2001. PMC 86928. PMID 11287605.
- ^ Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ (November 2000). "Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta". Genes & Development. 14 (22): 2819–2830. doi:10.1101/gad.844900. PMC 317055. PMID 11090130.
- ^ Na M, Jang J, Min BS, Lee SJ, Lee MS, Kim BY, Oh WK, Ahn JS (September 2006). "Fatty acid synthase inhibitory activity of acylphloroglucinols isolated from Dryopteris crassirhizoma". Bioorganic & Medicinal Chemistry Letters. 16 (18): 4738–4742. doi:10.1016/j.bmcl.2006.07.018. PMID 16870425.
- ^ Baron A, Migita T, Tang D, Loda M (January 2004). "Fatty acid synthase: a metabolic oncogene in prostate cancer?". Journal of Cellular Biochemistry. 91 (1): 47–53. doi:10.1002/jcb.10708. PMID 14689581. S2CID 26175683.
- ^ Hunt DA, Lane HM, Zygmont ME, Dervan PA, Hennigar RA (2007). "MRNA stability and overexpression of fatty acid synthase in human breast cancer cell lines". Anticancer Research. 27 (1A): 27–34. PMID 17352212.
- ^ Gansler TS, Hardman W, Hunt DA, Schaffel S, Hennigar RA (June 1997). "Increased expression of fatty acid synthase (OA-519) in ovarian neoplasms predicts shorter survival". Human Pathology. 28 (6): 686–692. doi:10.1016/S0046-8177(97)90177-5. PMID 9191002.
- ^ Ezzeddini R, Taghikhani M, Somi MH, Samadi N, Rasaee, MJ (May 2019). "Clinical importance of FASN in relation to HIF-1α and SREBP-1c in gastric adenocarcinoma". Life Sciences. 224: 169–176. doi:10.1016/j.lfs.2019.03.056. PMID 30914315. S2CID 85532042.
- ^ "First Human Study Taking Place With Fatty Acid Synthase Inhibitor". oncotherapynetwork.com. April 7, 2017. Archived from the original on April 15, 2019. Retrieved June 4, 2018.
- ^ Lu T, Schubert C, Cummings MD, Bignan G, Connolly PJ, Smans K, Ludovici D, Parker MH, Meyer C, Rocaboy C, Alexander R, Grasberger B, De Breucker S, Esser N, Fraiponts E, Gilissen R, Janssens B, Peeters D, Van Nuffel L, Vermeulen P, Bischoff J, Meerpoel L (May 2018). "Design and synthesis of a series of bioavailable fatty acid synthase (FASN) KR domain inhibitors for cancer therapy". Bioorganic & Medicinal Chemistry Letters. 28 (12): 2159–2164. doi:10.1016/j.bmcl.2018.05.014. PMID 29779975. S2CID 29159508.
- ^ Hardwicke MA, Rendina AR, Williams SP, Moore ML, Wang L, Krueger JA, Plant RN, Totoritis RD, Zhang G, Briand J, Burkhart WA, Brown KK, Parrish CA (September 2014). "A human fatty acid synthase inhibitor binds β-ketoacyl reductase in the keto-substrate site". Nature Chemical Biology. 10 (9): 774–779. doi:10.1038/nchembio.1603. PMID 25086508.
- ^ Vander Heiden MG, DeBerardinis RJ (February 2017). "Understanding the Intersections between Metabolism and Cancer Biology". Cell. 168 (4): 657–669. doi:10.1016/j.cell.2016.12.039. PMC 5329766. PMID 28187287.
- ^ Sgro CD (2009-01-01). An investigation into the interdomain region of Caenorhabditis elegans fatty acid synthase and its implications as a drug target (thesis thesis). La Trobe.
- ^ Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RR, Xu HE, Turk J, Semenkovich CF (August 2009). "Identification of a physiologically relevant endogenous ligand for PPARalpha in liver". Cell. 138 (3): 476–488. doi:10.1016/j.cell.2009.05.036. PMC 2725194. PMID 19646743.
- ^ Wu M, Singh SB, Wang J, Chung CC, Salituro G, Karanam BV, Lee SH, Powles M, Ellsworth KP, Lassman ME, Miller C, Myers RW, Tota MR, Zhang BB, Li C (March 2011). "Antidiabetic and antisteatotic effects of the selective fatty acid synthase (FAS) inhibitor platensimycin in mouse models of diabetes". Proceedings of the National Academy of Sciences of the United States of America. 108 (13): 5378–5383. Bibcode:2011PNAS..108.5378W. doi:10.1073/pnas.1002588108. PMC 3069196. PMID 21389266.
- ^ Flavin R, Peluso S, Nguyen PL, Loda M (April 2010). "Fatty acid synthase as a potential therapeutic target in cancer". Future Oncology. 6 (4): 551–562. doi:10.2217/fon.10.11. PMC 3197858. PMID 20373869.
- ^ Richardson RD, Ma G, Oyola Y, Zancanella M, Knowles LM, Cieplak P, Romo D, Smith JW (September 2008). "Synthesis of novel beta-lactone inhibitors of fatty acid synthase". Journal of Medicinal Chemistry. 51 (17): 5285–5296. doi:10.1021/jm800321h. PMC 3172131. PMID 18710210.
- ^ Eggert SL, Huyck KL, Somasundaram P, Kavalla R, Stewart EA, Lu AT, Painter JN, Montgomery GW, Medland SE, Nyholt DR, Treloar SA, Zondervan KT, Heath AC, Madden PA, Rose L, Buring JE, Ridker PM, Chasman DI, Martin NG, Cantor RM, Morton CC (2012). "Genome-wide linkage and association analyses implicate FASN in predisposition to uterine leiomyomata". American Journal of Human Genetics. 91 (4): 621–628. doi:10.1016/j.ajhg.2012.08.009. PMC 3484658. PMID 23040493.
Further reading
[edit]- Wakil SJ (1989). "Fatty acid synthase, a proficient multifunctional enzyme". Biochemistry. 28 (11): 4523–4530. doi:10.1021/bi00437a001. PMID 2669958.
- Baron A, Migita T, Tang D, Loda M (2004). "Fatty acid synthase: a metabolic oncogene in prostate cancer?". Journal of Cellular Biochemistry. 91 (1): 47–53. doi:10.1002/jcb.10708. PMID 14689581. S2CID 26175683.
- Lejin D (1978). "[Viscosimetry in clinical practice]". Medicinski Pregled. 30 (9–10): 477–482. PMID 600212.
- Wronkowski Z (1976). "[Cancer diagnosis of the respiratory system]". Pielȩgniarka I Połozna (12): 7–8. PMID 1044453.
- Semenkovich CF, Coleman T, Fiedorek FT (1995). "Human fatty acid synthase mRNA: tissue distribution, genetic mapping, and kinetics of decay after glucose deprivation". Journal of Lipid Research. 36 (7): 1507–1521. doi:10.1016/S0022-2275(20)39738-8. PMID 7595075.
- Kuhajda FP, Jenner K, Wood FD, Hennigar RA, Jacobs LB, Dick JD, Pasternack GR (1994). "Fatty acid synthesis: a potential selective target for antineoplastic therapy". Proceedings of the National Academy of Sciences of the United States of America. 91 (14): 6379–6383. Bibcode:1994PNAS...91.6379K. doi:10.1073/pnas.91.14.6379. PMC 44205. PMID 8022791.
- Hsu MH, Chirala SS, Wakil SJ (1996). "Human fatty-acid synthase gene. Evidence for the presence of two promoters and their functional interaction". Journal of Biological Chemistry. 271 (23): 13584–13592. doi:10.1074/jbc.271.23.13584. PMID 8662758.
- Pizer ES, Kurman RJ, Pasternack GR, Kuhajda FP (1997). "Expression of fatty acid synthase is closely linked to proliferation and stromal decidualization in cycling endometrium". International Journal of Gynecological Pathology. 16 (1): 45–51. doi:10.1097/00004347-199701000-00008. PMID 8986532. S2CID 45195801.
- Jayakumar A, Chirala SS, Wakil SJ (1997). "Human fatty acid synthase: assembling recombinant halves of the fatty acid synthase subunit protein reconstitutes enzyme activity". Proceedings of the National Academy of Sciences of the United States of America. 94 (23): 12326–12330. Bibcode:1997PNAS...9412326J. doi:10.1073/pnas.94.23.12326. PMC 24928. PMID 9356448.
- Kusakabe T, Maeda M, Hoshi N, Sugino T, Watanabe K, Fukuda T, Suzuki T (2000). "Fatty acid synthase is expressed mainly in adult hormone-sensitive cells or cells with high lipid metabolism and in proliferating fetal cells". Journal of Histochemistry and Cytochemistry. 48 (5): 613–622. doi:10.1177/002215540004800505. PMID 10769045.
- Ye Q, Chung LW, Li S, Zhau HE (2000). "Identification of a novel FAS/ER-alpha fusion transcript expressed in human cancer cells". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1493 (3): 373–377. doi:10.1016/s0167-4781(00)00202-5. PMID 11018265.
- Rochat-Steiner V, Becker K, Micheau O, Schneider P, Burns K, Tschopp J (2000). "FIST/HIPK3: a Fas/FADD-interacting serine/threonine kinase that induces FADD phosphorylation and inhibits fas-mediated Jun NH(2)-terminal kinase activation". Journal of Experimental Medicine. 192 (8): 1165–1174. doi:10.1084/jem.192.8.1165. PMC 2311455. PMID 11034606.
- Chirala SS, Jayakumar A, Gu ZW, Wakil SJ (2001). "Human fatty acid synthase: role of interdomain in the formation of catalytically active synthase dimer". Proceedings of the National Academy of Sciences of the United States of America. 98 (6): 3104–3108. Bibcode:2001PNAS...98.3104C. doi:10.1073/pnas.051635998. PMC 30614. PMID 11248039.
- Brink J, Ludtke SJ, Yang CY, Gu ZW, Wakil SJ, Chiu W (2002). "Quaternary structure of human fatty acid synthase by electron cryomicroscopy". Proceedings of the National Academy of Sciences of the United States of America. 99 (1): 138–143. Bibcode:2002PNAS...99..138B. doi:10.1073/pnas.012589499. PMC 117528. PMID 11756679.
- Joseph SB, Laffitte BA, Patel PH, Watson MA, Matsukuma KE, Walczak R, Collins JL, Osborne TF, Tontonoz P (2002). "Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors". Journal of Biological Chemistry. 277 (13): 11019–11025. doi:10.1074/jbc.M111041200. PMID 11790787.
- Ming D, Kong Y, Wakil SJ, Brink J, Ma J (2002). "Domain movements in human fatty acid synthase by quantized elastic deformational model". Proceedings of the National Academy of Sciences of the United States of America. 99 (12): 7895–7899. Bibcode:2002PNAS...99.7895M. doi:10.1073/pnas.112222299. PMC 122991. PMID 12060737.
- Field FJ, Born E, Murthy S, Mathur SN (2003). "Polyunsaturated fatty acids decrease the expression of sterol regulatory element-binding protein-1 in CaCo-2 cells: effect on fatty acid synthesis and triacylglycerol transport". Biochemical Journal. 368 (Pt 3): 855–864. doi:10.1042/BJ20020731. PMC 1223029. PMID 12213084.
External links
[edit]- Fatty+Acid+Synthase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Fatty Acid Synthesis: Rensselaer Polytechnic Institute
- Fatty Acid Synthase: RCSB PDB Molecule of the Month Archived 2014-07-14 at the Wayback Machine
- 3D electron microscopy structures of fatty acid synthase from the EM Data Bank(EMDB)
- PDBe-KB provides an overview of all the structure information available in the PDB for Human Fatty acid synthase