ELISA
| ELISA | |
|---|---|
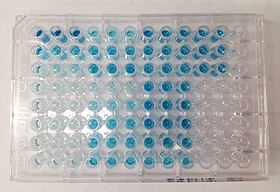 An ELISA being developed with TMB substrate for horseradish peroxidase-linked secondary antibody | |
| MeSH | D004797 |
The enzyme-linked immunosorbent assay (ELISA) (/ɪˈlaɪzə/, /ˌiːˈlaɪzə/) is a commonly used analytical biochemistry assay, first described by Eva Engvall and Peter Perlmann in 1971.[1] The assay is a solid-phase type of enzyme immunoassay (EIA) to detect the presence of a ligand (commonly a protein) in a liquid sample using antibodies directed against the ligand to be measured. ELISA has been used as a diagnostic tool in medicine, plant pathology, and biotechnology, as well as a quality control check in various industries.
In the most simple form of an ELISA, antigens from the sample to be tested are attached to a surface. Then, a matching antibody is applied over the surface so it can bind the antigen. This antibody is linked to an enzyme and then any unbound antibodies are removed. In the final step, a substance containing the enzyme's substrate is added. If there was binding, the subsequent reaction produces a detectable signal, most commonly a color change.
Performing an ELISA involves at least one antibody with specificity for a particular antigen. The sample with an unknown amount of antigen is immobilized on a solid support (usually a polystyrene microtiter plate) either non-specifically (via adsorption to the surface) or specifically (via capture by another antibody specific to the same antigen, in a "sandwich" ELISA). After the antigen is immobilized, the detection antibody is added, forming a complex with the antigen. The detection antibody can be covalently linked to an enzyme or can itself be detected by a secondary antibody that is linked to an enzyme through bioconjugation. Between each step, the plate is typically washed with a mild detergent solution to remove any proteins or antibodies that are non-specifically bound. After the final wash step, the plate is developed by adding an enzymatic substrate to produce a visible signal, which indicates the quantity of antigen in the sample.
Of note, ELISA can perform other forms of ligand binding assays instead of strictly "immuno" assays, though the name carried the original "immuno" because of the common use and history of development of this method. The technique essentially requires any ligating reagent that can be immobilized on the solid phase along with a detection reagent that will bind specifically and use an enzyme to generate a signal that can be properly quantified. In between the washes, only the ligand and its specific binding counterparts remain specifically bound or "immunosorbed" by antigen-antibody interactions to the solid phase, while the nonspecific or unbound components are washed away. Unlike other spectrophotometric wet lab assay formats where the same reaction well (e.g., a cuvette) can be reused after washing, the ELISA plates have the reaction products immunosorbed on the solid phase, which is part of the plate, and so are not easily reusable.
Principle
[edit]As an analytical biochemistry assay and a "wet lab" technique, ELISA involves detection of an analyte (i.e., the specific substance whose presence is being quantitatively or qualitatively analyzed) in a liquid sample by a method that continues to use liquid reagents during the analysis (i.e., controlled sequence of biochemical reactions that will generate a signal which can be easily quantified and interpreted as a measure of the amount of analyte in the sample) that stays liquid and remains inside a reaction chamber or well needed to keep the reactants contained.[2][3] This is in contrast to "dry lab" techniques that use dry strips. Even if the sample is liquid (e.g., a measured small drop), the final detection step in "dry" analysis involves reading of a dried strip by methods such as reflectometry and does not need a reaction containment chamber to prevent spillover or mixing between samples.[4]
As a heterogenous assay, ELISA separates some component of the analytical reaction mixture by adsorbing certain components onto a solid phase which is physically immobilized. In ELISA, a liquid sample is added onto a stationary solid phase with special binding properties and is followed by multiple liquid reagents that are sequentially added, incubated, and washed, followed by some optical change (e.g., color development by the product of an enzymatic reaction) in the final liquid in the well from which the quantity of the analyte is measured. The quantitative "reading" is usually based on detection of intensity of transmitted light by spectrophotometry, which involves quantitation of transmission of some specific wavelength of light through the liquid (as well as the transparent bottom of the well in the multiple-well plate format).[2][3] The sensitivity of detection depends on amplification of the signal during the analytic reactions. Since enzyme reactions are very well known amplification processes, the signal is generated by enzymes which are linked to the detection reagents in fixed proportions to allow accurate quantification, and thus the name "enzyme-linked".[5]
The analyte is also called the ligand because it will specifically bind or ligate to a detection reagent, thus ELISA falls under the bigger category of ligand binding assays.[2] The ligand-specific binding reagent is "immobilized", i.e., usually coated and dried onto the transparent bottom and sometimes also side wall of a well[6] (the stationary "solid phase"/"solid substrate" here as opposed to solid microparticle/beads that can be washed away), which is usually constructed as a multiple-well plate known as the "ELISA plate". Conventionally, like other forms of immunoassays, the specificity of antigen-antibody type reaction is used because it is easy to raise an antibody specifically against an antigen in bulk as a reagent. Alternatively, if the analyte itself is an antibody, its target antigen can be used as the binding reagent.[7]
History
[edit]Before the development of the ELISA, the only option for conducting an immunoassay was radioimmunoassay, a technique using radioactively labeled antigens or antibodies. In radioimmunoassay, the radioactivity provides the signal, which indicates whether a specific antigen or antibody is present in the sample. Radioimmunoassay was first described in a scientific paper by Rosalyn Sussman Yalow and Solomon Berson published in 1960.[8]
As radioactivity poses a potential health threat, a safer alternative was sought. A suitable alternative to radioimmunoassay would substitute a nonradioactive signal in place of the radioactive signal. When enzymes (such as horseradish peroxidase) react with appropriate substrates (such as ABTS or TMB), a change in color occurs, which is used as a signal. However, the signal has to be associated with the presence of antibody or antigen, which is why the enzyme has to be linked to an appropriate antibody. This linking process was independently developed by Stratis Avrameas and G. B. Pierce.[9] Since it is necessary to remove any unbound antibody or antigen by washing, the antibody or antigen has to be fixed to the surface of the container; i.e., the immunosorbent must be prepared. A technique to accomplish this was published by Wide and Jerker Porath in 1966.[10]
In 1971, Peter Perlmann and Eva Engvall at Stockholm University in Sweden, and Anton Schuurs and Bauke van Weemen in the Netherlands independently published papers that synthesized this knowledge into methods to perform EIA/ELISA.[11][12]
Traditional ELISA typically involves chromogenic reporters and substrates that produce some kind of observable color change to indicate the presence of antigen or analyte. Newer ELISA-like techniques use fluorogenic, electrochemiluminescent, and quantitative PCR reporters to create quantifiable signals. These new reporters can have various advantages, including higher sensitivities and multiplexing.[13][14] In technical terms, newer assays of this type are not strictly ELISAs, as they are not "enzyme-linked", but are instead linked to some nonenzymatic reporter. However, given that the general principles in these assays are largely similar, they are often grouped in the same category as ELISAs.
In 2012, an ultrasensitive, enzyme-based ELISA test using nanoparticles as a chromogenic reporter was able to give a naked-eye colour signal, from the detection of mere attograms of analyte. A blue color appears for positive results and red color for negative. Note that this detection only can confirm the presence or the absence of analyte, not the actual concentration.[15]
Types
[edit]There are many ELISA tests for particular molecules that use the matching antibodies. ELISA tests are broken into several types of tests based on how the analytes and antibodies are bonded and used.[16][17] The major types are described here.[18]
Direct
[edit]
The steps of direct ELISA[19] follows the mechanism below:
- A buffered solution of the antigen to be tested for is added to each well (usually 96-well plates) of a microtiter plate, where it is given time to adhere to the plastic through charge interactions.
- A solution of nonreacting protein, such as bovine serum albumin or casein, is added to each well in order to cover any plastic surface in the well which remains uncoated by the antigen.
- The primary antibody with an attached (conjugated) enzyme is added, which binds specifically to the test antigen coating the well.
- A substrate for this enzyme is then added. Often, this substrate changes color upon reaction with the enzyme.
- The higher the concentration of the primary antibody present in the serum, the stronger the color change. Often, a spectrometer is used to give quantitative values for color strength.
The enzyme acts as an amplifier; even if only few enzyme-linked antibodies remain bound, the enzyme molecules will produce many signal molecules. Within common-sense limitations, the enzyme can go on producing color indefinitely, but the more antibody is bound, the faster the color will develop. A major disadvantage of the direct ELISA is that the method of antigen immobilization is not specific; when serum is used as the source of test antigen, all proteins in the sample may stick to the microtiter plate well, so small concentrations of analyte in serum must compete with other serum proteins when binding to the well surface. The sandwich or indirect ELISA provides a solution to this problem, by using a "capture" antibody specific for the test antigen to pull it out of the serum's molecular mixture.[citation needed]
ELISA may be run in a qualitative or quantitative format. Qualitative results provide a simple positive or negative result (yes or no) for a sample. The cutoff between positive and negative is determined by the analyst and may be statistical. Two or three times the standard deviation (error inherent in a test) is often used to distinguish positive from negative samples. In quantitative ELISA, the optical density (OD) of the sample is compared to a standard curve, which is typically a serial dilution of a known-concentration solution of the target molecule. For example, if a test sample returns an OD of 1.0, the point on the standard curve that gave OD = 1.0 must be of the same analyte concentration as the sample.[citation needed]
The use and meaning of the names "indirect ELISA" and "direct ELISA" differs in the literature and on web sites depending on the context of the experiment. When the presence of an antigen is analyzed, the name "direct ELISA" refers to an ELISA in which only a labelled primary antibody is used, and the term "indirect ELISA" refers to an ELISA in which the antigen is bound by the primary antibody which then is detected by a labeled secondary antibody. In the latter case a sandwich ELISA is clearly distinct from an indirect ELISA. When the "primary" antibody is of interest, e.g. in the case of immunization analyses, this antibody is directly detected by the secondary antibody and the term "indirect ELISA" applies to a setting with two antibodies.[citation needed]
Sandwich
[edit]
A "sandwich" ELISA is used to detect sample antigen.[20] The steps are:
- A surface is prepared with a known quantity of capture antibody.
- Any nonspecific binding sites on the surface are blocked.
- The antigen-containing sample is applied to the plate, and captured by antibody.
- The plate is washed to remove unbound antigen.
- A specific antibody is added, and binds to antigen (hence the 'sandwich': the antigen is stuck between two antibodies). This primary antibody could be in the serum of a donor, to be tested for reactivity towards the antigen.
- Enzyme-linked secondary antibodies are applied as detection antibodies, which bind specifically to the antibody's Fc region (nonspecific).
- The plate is washed to remove the unbound antibody-enzyme conjugates.
- A chemical is added to be converted by the enzyme into a color, fluorescent, or electrochemical signal.
- The absorbance, fluorescence, or electrochemical signal (e.g., current) of the plate's wells is measured to determine the presence and quantity of the antigen.
The image to the right includes the use of a secondary antibody conjugated to an enzyme, although, in the technical sense, this is not necessary if the primary antibody is conjugated to an enzyme (which would be direct ELISA). However, the use of a secondary-antibody conjugate avoids the expensive process of creating enzyme-linked antibodies for every antigen one might want to detect. By using an enzyme-linked antibody that binds the Fc region of other antibodies, this same enzyme-linked antibody can be used in a variety of situations. Without the first layer of "capture" antibody, any proteins in the sample (including serum proteins) may competitively adsorb to the plate surface, lowering the quantity of antigen immobilized. Use of the purified specific antibody to attach the antigen to the plastic eliminates a need to purify the antigen from complicated mixtures before the measurement, simplifying the assay, and increasing the specificity and the sensitivity of the assay. Therefore, a sandwich ELISA used for research often needs validation, to reduce the risk of false positive results.[21]
Competitive
[edit]A third use of ELISA is through competitive binding. The steps for this ELISA are somewhat different from the first two examples:
Unlabeled antibody is incubated in the presence of its antigen (sample).
- These bound antibody/antigen complexes are then added to an antigen-coated well.
- The plate is washed, so unbound antibodies are removed. (The more antigen in the sample, the more Ag-Ab complexes are formed and so there are less unbound antibodies available to bind to the antigen in the well, hence "competition".)
- The secondary antibody, specific to the primary antibody, is added. This second antibody is coupled to the enzyme.
- A substrate is added, and remaining enzymes elicit a chromogenic or fluorescent signal.
- The reaction is stopped to prevent eventual saturation of the signal.
Some competitive ELISA kits include enzyme-linked antigen rather than enzyme-linked antibody. The labeled antigen competes for primary antibody binding sites with the sample antigen (unlabeled). The less antigen in the sample, the more labeled antigen is retained in the well and the stronger the signal.
Commonly, the antigen is not first positioned in the well.
For the detection of HIV antibodies, the wells of microtiter plate are coated with the HIV antigen. Two specific antibodies are used, one conjugated with enzyme and the other present in serum (if serum is positive for the antibody). Cumulative competition occurs between the two antibodies for the same antigen, causing a stronger signal to be seen. Sera to be tested are added to these wells and incubated at 37 °C, and then washed. If antibodies are present, the antigen-antibody reaction occurs. No antigen is left for the enzyme-labelled specific HIV antibodies. These antibodies remain free upon addition and are washed off during washing. Substrate is added, but there is no enzyme to act on it, so a positive result shows no color change.
Indirect
[edit]A fourth ELISA test does not use the traditional wells, rather leaves the antigens suspended in the test fluid.[22][23]
- Unlabeled antibody is incubated in the presence of its antigen (sample)
- A sufficient incubation period is provided to allow the antibodies to bind to the antigens.
- The sample is then passed through the Scavenger container. This can be a test tube or a specifically designed flow through channel. The surface of the Scavenger container or channel has "Scavenger Antigens" bound to it. These can be identical or sufficiently similar to the primary antigens that the free antibodies will bind.
- The Scavenger container must have sufficient surface area and sufficient time to allow the Scavenger Antigens to bind to all the excess Antibodies introduced into the sample.
- The sample, that now contains the tagged and bound antibodies, is passed through a detector. This device can be a flow cytometer or other device that illuminates the tags and registers the response.[24]
This test allows multiple antigens to be tagged and counted at the same time. This allows specific strains of bacteria to be identified by two (or more) different color tags. If both tags are present on a cell, then the cell is that specific strain. If only one is present, it is not.
This test is done, generally, one test at a time and cannot be done with the microtiter plate. The equipment needed is usually less complicated and can be used in the field.
Commonly used enzymatic markers
[edit]The following table lists the enzymatic markers commonly used in ELISA assays, which allow the results of the assay to be measured upon completion.
- OPD (o-phenylenediamine dihydrochloride) turns amber to detect HRP (horseradish peroxidase), which is often used to as a conjugated protein.[25]
- TMB (3,3',5,5'-tetramethylbenzidine) turns blue when detecting HRP and turns yellow after the addition of sulfuric or phosphoric acid.[25]
- ABTS (2,2'-Azinobis [3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt) turns green when detecting HRP.[25]
- PNPP (p-Nitrophenyl Phosphate, Disodium Salt) turns yellow when detecting alkaline phosphatase.[25]
Applications
[edit]
Because the ELISA can be performed to evaluate either the presence of antigen or the presence of antibody in a sample, it is a useful tool for determining serum antibody concentrations (such as with the HIV test[26] or West Nile virus). It has also found applications in the food industry in detecting potential food allergens, such as milk, peanuts, walnuts, almonds, and eggs[27] and as serological blood test for coeliac disease.[28][29] ELISA can also be used in toxicology as a rapid presumptive screen for certain classes of drugs.

The ELISA was the first screening test widely used for HIV because of its high sensitivity. In an ELISA, a person's serum is diluted 400 times and applied to a plate to which HIV antigens are attached. If antibodies to HIV are present in the serum, they may bind to these HIV antigens. The plate is then washed to remove all other components of the serum. A specially prepared "secondary antibody"—an antibody that binds to other antibodies—is then applied to the plate, followed by another wash. This secondary antibody is chemically linked in advance to an enzyme.
Thus, the plate will contain enzyme in proportion to the amount of secondary antibody bound to the plate. A substrate for the enzyme is applied, and catalysis by the enzyme leads to a change in color or fluorescence. ELISA results are reported as a number; the most controversial aspect of this test is determining the "cut-off" point between a positive and a negative result.
A cut-off point may be determined by comparing it with a known standard. If an ELISA test is used for drug screening at workplace, a cut-off concentration, 50 ng/ml, for example, is established, and a sample containing the standard concentration of analyte will be prepared. Unknowns that generate a stronger signal than the known sample are "positive". Those that generate weaker signal are "negative".
There are ELISA tests to detect various kind of diseases, such as dengue, malaria, Chagas disease,[30] Johne's disease, and others.[31] ELISA tests also are extensively employed for in vitro diagnostics in medical laboratories. The other uses of ELISA include:
- detection of SARS-CoV-2 antibodies in blood samples[32]

ELISA is as of 2023 the primary method of plant pathogen detection worldwide.[33]
Enzyme-Linked Single Molecule Array (eSimoa)
[edit]eSimoa (enzyme-linked single molecule array) represents a significant evolution of the traditional ELISA (Enzyme-Linked Immunosorbent Assay) technique, which is widely utilized in clinical diagnostics and research. By significantly enhancing the sensitivity and resolution of biomolecular detection, eSimoa expands the capabilities of ELISA, enabling the detection of biomolecules at concentrations previously unachievable with standard assays.[34]
Technology
[edit]Building on the foundational principles of ELISA, eSimoa employs paramagnetic beads to isolate biomolecules or enzymes in a manner akin to ELISA’s plate-based detection. However, eSimoa advances this concept by enabling enzymatic reaction measurements at the single-molecule level, which dramatically improves detection limits for various enzymes and biomolecules. This method allows for the precise quantification of low-abundance proteins and the activity of critical enzymes such as protein kinases and telomerases, which are often below the detection threshold of conventional ELISA.
Applications
[edit]The enhanced sensitivity of eSimoa is crucial for early and accurate biomarker detection in clinical diagnostics, facilitating better disease monitoring and management. In drug discovery, the ability to track subtle changes in enzymatic activity aids in the development of more effective pharmaceuticals by providing detailed insights into enzyme inhibition mechanisms.
Origins and Controversy
[edit]Chi-An Cheng at National Taiwan University (NTU) has claimed that her team developed this innovative technology.[35][36] However, this claim is contested by the existence of prior publications by David R. Walt's team at Harvard University, who published their work on eSimoa in 2020.[34][37] This earlier documentation by Walt's team suggests a prior contribution to the development of the technology.
See also
[edit]- Agglutination-PCR
- Immunoscreening
- Lateral flow test
- Magnetic immunoassay
- Microtitre plate
- Nephelometry
- Plaque reduction neutralization test
- Plate reader
- Secretion assay
Notes and references
[edit]- ^ Engvall, E (1972-11-22). "Enzyme-linked immunosorbent assay, Elisa". The Journal of Immunology. 109 (1): 129–135. doi:10.4049/jimmunol.109.1.129. ISSN 0022-1767. PMID 4113792.
- ^ a b c Msagati, T.A. (2017). Food Forensics and Toxicology. John Wiley & Sons. p. PT229. ISBN 9781119101383.
- ^ a b Crowther, J.R. (1995). "Chapter 2: Basic Principles of ELISA". ELISA: Theory and Practice. Methods in Molecular Biology. Vol. 42. Humana Press. pp. 35–62. doi:10.1385/0-89603-279-5:1. ISBN 0896032795. PMID 7655571.
- ^ Sonntag, O. (1993). "Chapter 1: Introduction to dry chemistry". In van der Vliet, P.C. (ed.). Dry Chemistry: Analysis with carrier-bound reagents. Laboratory Techniques in Biochemistry and Molecular Biology. Vol. 25. pp. 1–6. ISBN 9780080887364.
- ^ Hsieh, Y.-H.P.; Rao, Q. (2017). Nielsen, S.S. (ed.). Food Analysis. Springer. pp. 491–98. ISBN 9783319457765.
- ^ Schasfoort, R.B.M. (2017). Handbook of Surface Plasmon Resonance (2nd ed.). Royal Society of Chemistry. p. 296. ISBN 9781782627302.
- ^ Elgert, K.D. (2009). Immunology: Understanding the Immune System. John Wiley & Sons. pp. 149–50. ISBN 9780470081570.
- ^ Yalow, Rosalyn S.; Berson, Solomon A. (1960). "Immunoassay of endogenous plasma insulin in man". The Journal of Clinical Investigation. 39 (7): 1157–75. doi:10.1172/JCI104130. PMC 441860. PMID 13846364.
- ^ Lequin, R. M. (2005). "Enzyme Immunoassay (EIA)/Enzyme-Linked Immunosorbent Assay (ELISA)". Clinical Chemistry. 51 (12): 2415–8. doi:10.1373/clinchem.2005.051532. PMID 16179424.
- ^ Wide, Leif; Porath, Jerker (1966). "Radioimmunoassay of proteins with the use of Sephadex-coupled antibodies". Biochimica et Biophysica Acta (BBA) - General Subjects. 130 (1): 257–60. doi:10.1016/0304-4165(66)90032-8.
- ^ Engvall, Eva; Perlmann, Peter (1971). "Enzyme-linked immunosorbent assay (ELISA) quantitative assay of immunoglobulin G". Immunochemistry. 8 (9): 871–4. doi:10.1016/0019-2791(71)90454-X. PMID 5135623.
- ^ Van Weemen, B.K.; Schuurs, A.H.W.M. (1971). "Immunoassay using antigen—enzyme conjugates". FEBS Letters. 15 (3): 232–236. doi:10.1016/0014-5793(71)80319-8. PMID 11945853. S2CID 37147723.
- ^ Leng, S. X.; McElhaney, J. E.; Walston, J. D.; Xie, D.; Fedarko, N. S.; Kuchel, G. A. (2008). "ELISA and Multiplex Technologies for Cytokine Measurement in Inflammation and Aging Research". The Journals of Gerontology Series A: Biological Sciences and Medical Sciences. 63 (8): 879–84. doi:10.1093/gerona/63.8.879. PMC 2562869. PMID 18772478.
- ^ Adler, Michael; Schulz, Sven; Spengler, Mark (2009). "Cytokine Quantification in Drug Development: A comparison of sensitive immunoassay platforms". Chimera Biotech. Archived from the original on 2015-12-22. Retrieved 2015-08-20.
- ^ de la Rica, Roberto; Stevens, Molly M. (2012). "Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye". Nature Nanotechnology. 7 (12): 821–4. Bibcode:2012NatNa...7..821D. doi:10.1038/nnano.2012.186. hdl:10044/1/21938. PMID 23103935.
- ^ R., Crowther, J. (1995). "Basic Immunology". ELISA : theory and practice. Methods in Molecular Biology. Vol. 42. Totowa, N.J.: Humana Press. pp. 1–218. doi:10.1385/0-89603-279-5:1. ISBN 978-0896032798. OCLC 32130600. PMID 7655571.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ ROBERT., HNASKO (2016). ELISA (SOFTCOVER reprint OF ed.). [Place of publication not identified]: HUMANA. ISBN 978-1493953851. OCLC 960834982.
- ^ "What is an ELISA?". R&D Systems. Retrieved 31 January 2020.
- ^ Spence, Zachary (2018-10-18). "Biochemistry 8th ed - Jeremy M. Berg": 83.
{{cite journal}}: Cite journal requires|journal=(help)[permanent dead link] - ^ Schmidt, SD; Mazzella, MJ; Nixon, RA; Mathews, PM (2012). "Aβ Measurement by Enzyme-Linked Immunosorbent Assay". Amyloid Proteins. Methods in Molecular Biology. Vol. 849. pp. 507–27. doi:10.1007/978-1-61779-551-0_34. ISBN 978-1-61779-550-3. PMID 22528112.
- ^ Kragstrup, Tue W; Vorup-Jensen, Thomas; Deleuran, Bent; Hvid, Malene (2013). "A simple set of validation steps identifies and removes false results in a sandwich enzyme-linked immunosorbent assay caused by anti-animal IgG antibodies in plasma from arthritis patients". SpringerPlus. 2 (1): 263. doi:10.1186/2193-1801-2-263. PMC 3695686. PMID 23875127.
- ^ US 7767404, Charbonnet, Derrick, "Apparatus and method for single-step immunosorbent assay for single and multiple analytes", issued August 3, 2010
- ^ US 8735142, Charbonnet, Derrick & Evans, Norman Scott, "Systems and methods for immunosorbent assays for single and multiple analytes", issued May 27, 2014
- ^ Mahmoudi Gomari, Mohammad; Saraygord-Afshari, Neda; Farsimadan, Marziye; Rostami, Neda; Aghamiri, Shahin; Farajollahi, Mohammad M. (December 2020). "Opportunities and challenges of the tag-assisted protein purification techniques: Applications in the pharmaceutical industry". Biotechnology Advances. 45: 107653. doi:10.1016/j.biotechadv.2020.107653. ISSN 0734-9750. PMID 33157154. S2CID 226276355.
- ^ a b c d "Enzyme Substrates for ELISA". Thermo Fisher Scientific - US. Retrieved 2018-06-06.
- ^ MedlinePlus Encyclopedia: ELISA/Western blot tests for HIV
- ^ "Food Allergen Partnership" (Press release). FDA. January 2001. Retrieved August 20, 2015.
- ^ Sblattero, D.; Berti, I.; Trevisiol, C.; Marzari, R.; Tommasini, A.; Bradbury, A.; Fasano, A.; Ventura, A.; Not, T. (2000). "Human recombinant tissue transglutaminase ELISA: an innovative diagnostic assay for celiac disease". The American Journal of Gastroenterology. 95 (5): 1253–7. doi:10.1111/j.1572-0241.2000.02018.x. PMID 10811336. S2CID 11018740.
- ^ Porcelli, Brunetta; Ferretti, Fabio; Vindigni, Carla; Terzuoli, Lucia (2014). "Assessment of a Test for the Screening and Diagnosis of Celiac Disease". Journal of Clinical Laboratory Analysis. 30 (1): 65–70. doi:10.1002/jcla.21816. PMC 6807240. PMID 25385391.
- ^ Del-Rei, Rodrigo Pimenta; Leony, Leonardo Maia; Celedon, Paola Alejandra Fiorani; Zanchin, Nilson Ivo Tonin; Reis, Mitermayer Galvão dos; Gomes, Yara de Miranda; Schijman, Alejandro Gabriel; Longhi, Silvia Andrea; Santos, Fred Luciano Neves (2019-04-18). "Detection of anti-Trypanosoma cruzi antibodies by chimeric antigens in chronic Chagas disease-individuals from endemic South American countries". PLOS ONE. 14 (4): e0215623. Bibcode:2019PLoSO..1415623D. doi:10.1371/journal.pone.0215623. ISSN 1932-6203. PMC 6472793. PMID 30998741.
- ^ Griffin, J. F. T.; Spittle, E.; Rodgers, C. R.; Liggett, S.; Cooper, M.; Bakker, D.; Bannantine, J. P. (2005). "Immunoglobulin G1 Enzyme-Linked Immunosorbent Assay for Diagnosis of Johne's Disease in Red Deer (Cervus elaphus)". Clinical and Vaccine Immunology. 12 (12): 1401–9. doi:10.1128/CDLI.12.12.1401-1409.2005. PMC 1317074. PMID 16339063.
- ^ Dhamad, AE; Abdal Rhida, MA (2020). "COVID-19: molecular and serological detection methods". PeerJ. 8: e10180. doi:10.7717/peerj.10180. PMC 7547594. PMID 33083156.
- ^ Current and emerging trends in techniques for plant pathogen detection Frontiers in Plant Science
- ^ a b Wang, Xu; Ogata, Alana F.; Walt, David R. (2 September 2020). "Ultrasensitive Detection of Enzymatic Activity Using Single Molecule Arrays". Journal of the American Chemical Society. 142 (35): 15098–15106. doi:10.1021/jacs.0c06599. PMC 7472518. PMID 32797755.
- ^ Cheng, CA (28 November 2023). "eSimoa – ultrasensitive and high-resolution protein spatially decoding framework for tumor extracellular vesicles | Exosome RNA". Exosome RNA.
- ^ Cheng, Chi-An (13 June 2024). "Voices in Molecular Pharmaceutics : Meet Professor Chi-An Cheng, Who Is Innovating Diagnostic Tools, Discovering Biomarkers, Developing New Assays, and Creating New Materials". Molecular Pharmaceutics. doi:10.1021/acs.molpharmaceut.4c00623. PMID 38869420.
- ^ "Detection of Other Biomolecules – Walt Lab".
External links
[edit]- ELISA at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- "Introduction to ELISA Activity"—beginner walk-through of ELISA used for detecting HIV, including animations at the University of Arizona
- "ELISA Assay Principle" - An overview of the principle of an ELISA and tools for detecting analyties at Assay Genie
