Cystathionine beta-lyase
| cystathionine beta-lyase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
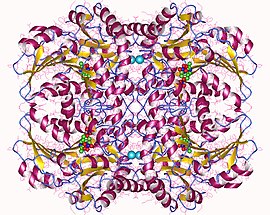 Cystathionine beta-lyase tetramer, E.Coli | |||||||||
| Identifiers | |||||||||
| EC no. | 4.4.1.8 | ||||||||
| CAS no. | 9055-05-4 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Cystathionine beta-lyase (EC 4.4.1.8), also commonly referred to as CBL or β-cystathionase, is an enzyme that primarily catalyzes the following α,β-elimination reaction[1]

Thus, the substrate of this enzyme is L-cystathionine, whereas its 3 products are homocysteine, pyruvate, and ammonia.[2][3][4]
Found in plants, bacteria, and yeast, cystathionine beta-lyase is an essential part of the methionine biosynthesis pathway as homocysteine can be directly converted into methionine by methionine synthase.[3][5][6] The enzyme belongs to the γ-family of PLP-dependent enzymes due to its use of a pyridoxal-5'-phosphate (PLP) cofactor to cleave cystathionine.[7] The enzyme also belongs to the family of lyases, specifically the class of carbon-sulfur lyases. The systematic name of this enzyme class is L-cystathionine L-homocysteine-lyase (deaminating; pyruvate-forming). This enzyme participates in 5 metabolic pathways: methionine metabolism, cysteine metabolism, selenoamino acid metabolism, nitrogen metabolism, and sulfur metabolism.
Structure
[edit]Cystathionine beta-lyase is a tetramer composed of identical subunits, and is constructed as a dimer of dimers, each associated with one molecule of PLP bound to the catalytic site by a lysine residue.[6][8] The dimer is formed by two monomers associated through several electrostatic, hydrogen bonding, and hydrophobic interactions, whereas the tetramer is stabilized through interactions between the N-terminal domains and key α-helices.[3]
Most of the enzyme's catalytic site residues are conserved amongst the enzymes involved in the transsulfuration pathway.[6] Other members include cystathionine gamma-synthase, cystathionine gamma-lyase, and methionine gamma lyase.[9][10] Additionally, these structures exhibit a type I fold and belong to the aspartate aminotransferase (AAT) family, characterized by homodimers with dihedral symmetry and active sites composed of residues belonging to adjacent subunits.[11][12]

Monomer
[edit]The cystathionine beta-lyase monomer consists of three functionally and structurally distinct domains:
N-terminal domain
[edit]Composed of three α-helices and one beta-strand that contribute to the formation of the quaternary structure.[6][13] This domain contains residues that interact with the active site of the neighboring subunit to facilitate substrate and cofactor binding.[4]
PLP-binding domain
[edit]Contains most of the catalytically relevant residues on the enzyme. It is composed of α-helices and β-sheets with a distinct parallel seven-stranded β-sheet. These sheets form a curved structure around the PLP-binding helix. PLP is covalently attached to a lysine residue at the C-terminus of the sheet.[3][4]
C-terminal domain
[edit]Smallest domain on the enzyme, which is attached to the PLP-binding domain by a long, kinked α-helix. The domain is structured into four-stranded antiparallel β-sheet with neighboring helices.[4]
Catalytic site
[edit]Aside from being bound to a lysine residue, PLP is fixed within the substrate binding site of the enzyme through various interactions with catalytic residues. Amine- and hydroxyl-containing residues are located in hydrogen bonding distance to the four phosphate oxygens.[3] This phosphate group is considered to be the main contributor to securing PLP in the active site. Additionally, residues neighboring the pyridine nitrogen in PLP help stabilize its positive charge, thereby increasing its electrophilic character.[14]
The aromatic ring in PLP is fixed in place by an almost coplanar tyrosine residue. It is believed that this configuration increases the electron sink character of the cofactor. These stacking interactions between PLP and aromatic side chains can be found in most PLP-dependent enzymes as it plays an important role in catalyzing the reaction by facilitating transaldimination.[15]

Mechanism
[edit]As shown in the mechanism below, cystathionine beta-lyase facilitates the S-C bond cleavage in cystathionine with the use of a PLP cofactor bounded to a catalytic lysine residue.[3][4] Initially, a deprotonated amino group is needed to perform the transaldimination reaction.[13] Given that the pH optimum for the enzyme is between 8.0 and 9.0, a tyrosine residue in the catalytic pocket exists as a phenolate, which abstracts a proton from the α-amino group of the substrate.[5][6] In the next step, the deprotonated amine undergoes a nucleophilic attack and displaces the lysine to form a Schiff base, forming an internal aldimine.
The released lysine can now abstract the proton from the Cα and form a quinoid intermediate, which is facilitated by the delocalization of the negative charge over PLP's conjugated p-system.[14] Subsequently, the protonation of Sγ induces Cβ-Sγ bond cleavage, thereby releasing homocysteine[3][13]
The external aldimine is displaced by the nucleophilic attack of the lysine, regenerating the catalytically active internal aldimine and releasing dehydroalanine.[4] Lastly, the enamine tautomerizes into an imine that undergoes hydrolytic deamination to form pyruvate and ammonia.[16]

Inhibition
[edit]Plant and bacterial cystathionine beta-lyases are inhibited by the antimicrobial amino acid, L-aminoethoxyvinylglycine (AVG), and the antibacterial amino acid, rhizobitoxine.[3]
Plants
[edit]Cystathionine beta-lyase in plants exhibits a two-step mechanism inactivation process with AVG, in which a reversible enzyme-inhibitor complex is formed before the irreversible inactivation of the enzyme:

Excess addition of cystathionine prevented the inactivation of the enzyme, suggesting that AVG acts as a competitive inhibitor with respect to cystathionine.[5] Additionally, the enzyme has been shown to be sensitive to thiol-blocking inhibitors, such as N-ethylmaleimide and iodoacetamide.[8][17]
Bacteria
[edit]Unlike in plants, Cystathionine beta-lyase in bacteria exhibits a one-step inhibition mechanism:

Through kinetic methods and X-ray crystallography, a time-dependent, slow-binding inhibition was observed. It is believed that the inhibitor binds to the enzyme in a similar way as the substrate; however, after the abstraction of the α-proton, the reaction proceeds to create an inactive ketimine PLP derivative.[18]

Evolution
[edit]Arabidopsis cystathionine beta-lyase possesses 22% homology with its Escherichia coli counterpart and even higher homology (between 28% and 36%) with cystathionine γ-synthase from plant and bacterial sources and cystathionine γ-lyase from Saccharomyces cerevisiae.[19] All of these enzymes are involved in the Cys/Met biosynthetic pathway and belong to the same class of PLP-dependent enzymes, suggesting that these enzymes were derived from a common ancestor.[6][20]
Industrial relevance
[edit]Cystathionine beta-lyase catalyzes the production of homocysteine, a direct precursor to methionine. Methionine is an essential amino acid for bacteria that is required for protein synthesis and the synthesis of S-adenosylmethionine; thus, the amino acid is directly linked to DNA replication. Because of its necessity in DNA replication, inhibition of cystathionine beta-lyase is an attractive antibiotic target.[21] Furthermore, the enzyme is absent in humans, decreasing the chance of harmful and unwanted side effects.[22]
Studies have linked the anti-fungal activity of several anti-fungal agents to the inhibition of cystathionine beta-lyase; however, other studies have not observed enzyme inhibition by these. Further research is needed to characterize the full extent cystathionine beta-lyase inhibition has on microbial and fungal growth.[21]
References
[edit]- ^ Dwivedi CM, Ragin RC, Uren JR (June 1982). "Cloning, purification, and characterization of beta-cystathionase from Escherichia coli". Biochemistry. 21 (13): 3064–9. doi:10.1021/bi00256a005. PMID 7049234.
- ^ Flavin M, Slaughter C (July 1964). "Cystathionine Cleavage Enzymes of Neurospora". The Journal of Biological Chemistry. 239 (7): 2212–9. doi:10.1016/S0021-9258(20)82222-4. PMID 14209950.
- ^ a b c d e f g h Breitinger U, Clausen T, Ehlert S, Huber R, Laber B, Schmidt F, Pohl E, Messerschmidt A (June 2001). "The three-dimensional structure of cystathionine beta-lyase from Arabidopsis and its substrate specificity". Plant Physiology. 126 (2): 631–42. doi:10.1104/pp.126.2.631. PMC 111155. PMID 11402193.
- ^ a b c d e f Clausen T, Laber B, Messerschmidt A (1997-03-01). "Mode of action of cystathionine beta-lyase". Biological Chemistry. 378 (3–4): 321–6. PMID 9165088.
- ^ a b c Droux M, Ravanel S, Douce R (January 1995). "Methionine biosynthesis in higher plants. II. Purification and characterization of cystathionine beta-lyase from spinach chloroplasts". Archives of Biochemistry and Biophysics. 316 (1): 585–95. doi:10.1006/abbi.1995.1078. PMID 7840670.
- ^ a b c d e f Messerschmidt A, Worbs M, Steegborn C, Wahl MC, Huber R, Laber B, Clausen T (March 2003). "Determinants of enzymatic specificity in the Cys-Met-metabolism PLP-dependent enzymes family: crystal structure of cystathionine gamma-lyase from yeast and intrafamiliar structure comparison". Biological Chemistry. 384 (3): 373–86. doi:10.1515/BC.2003.043. PMID 12715888. S2CID 24552794.
- ^ Alexander FW, Sandmeier E, Mehta PK, Christen P (February 1994). "Evolutionary relationships among pyridoxal-5'-phosphate-dependent enzymes. Regio-specific alpha, beta and gamma families". European Journal of Biochemistry. 219 (3): 953–60. doi:10.1111/j.1432-1033.1994.tb18577.x. PMID 8112347.
- ^ a b Ravanel S, Job D, Douce R (December 1996). "Purification and properties of cystathionine beta-lyase from Arabidopsis thaliana overexpressed in Escherichia coli". The Biochemical Journal. 320 ( Pt 2) (2): 383–92. doi:10.1042/bj3200383. PMC 1217943. PMID 8973544.
- ^ Holbrook EL, Greene RC, Krueger JH (January 1990). "Purification and properties of cystathionine gamma-synthase from overproducing strains of Escherichia coli". Biochemistry. 29 (2): 435–42. doi:10.1021/bi00454a019. PMID 2405903.
- ^ Kreft BD, Townsend A, Pohlenz HD, Laber B (April 1994). "Purification and Properties of Cystathionine [gamma]-Synthase from Wheat (Triticum aestivum L.)". Plant Physiology. 104 (4): 1215–1220. doi:10.1104/pp.104.4.1215. PMC 159283. PMID 12232160.
- ^ Grishin NV, Phillips MA, Goldsmith EJ (July 1995). "Modeling of the spatial structure of eukaryotic ornithine decarboxylases". Protein Science. 4 (7): 1291–304. doi:10.1002/pro.5560040705. PMC 2143167. PMID 7670372.
- ^ Jansonius, JN (December 1998). "Structure, evolution and action of vitamin B6-dependent enzymes". Current Opinion in Structural Biology. 8 (6): 759–69. doi:10.1016/s0959-440x(98)80096-1. PMID 9914259.
- ^ a b c Clausen T, Huber R, Laber B, Pohlenz HD, Messerschmidt A (September 1996). "Crystal structure of the pyridoxal-5'-phosphate dependent cystathionine beta-lyase from Escherichia coli at 1.83 A". Journal of Molecular Biology. 262 (2): 202–24. doi:10.1006/jmbi.1996.0508. PMID 8831789.
- ^ a b John RA (April 1995). "Pyridoxal phosphate-dependent enzymes". Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1248 (2): 81–96. doi:10.1016/0167-4838(95)00025-p. PMID 7748903.
- ^ Aitken SM, Lodha PH, Morneau DJ (November 2011). "The enzymes of the transsulfuration pathways: active-site characterizations". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 1814 (11): 1511–7. doi:10.1016/j.bbapap.2011.03.006. PMID 21435402.
- ^ "ENZYME entry 4.4.1.8". enzyme.expasy.org. Retrieved 2017-03-09.
- ^ Gentry-Weeks CR, Spokes J, Thompson J (March 1995). "beta-Cystathionase from Bordetella avium. Role(s) of lysine 214 and cysteine residues in activity and cytotoxicity". The Journal of Biological Chemistry. 270 (13): 7695–702. doi:10.1074/jbc.270.13.7695. PMID 7706318.
- ^ Clausen T, Huber R, Messerschmidt A, Pohlenz HD, Laber B (October 1997). "Slow-binding inhibition of Escherichia coli cystathionine beta-lyase by L-aminoethoxyvinylglycine: a kinetic and X-ray study". Biochemistry. 36 (41): 12633–43. doi:10.1021/bi970630m. PMID 9376370.
- ^ Ravanel S, Gakière B, Job D, Douce R (June 1998). "The specific features of methionine biosynthesis and metabolism in plants". Proceedings of the National Academy of Sciences of the United States of America. 95 (13): 7805–12. Bibcode:1998PNAS...95.7805R. doi:10.1073/pnas.95.13.7805. PMC 22764. PMID 9636232.
- ^ Belfaiza J, Parsot C, Martel A, de la Tour CB, Margarita D, Cohen GN, Saint-Girons I (February 1986). "Evolution in biosynthetic pathways: two enzymes catalyzing consecutive steps in methionine biosynthesis originate from a common ancestor and possess a similar regulatory region". Proceedings of the National Academy of Sciences of the United States of America. 83 (4): 867–71. Bibcode:1986PNAS...83..867B. doi:10.1073/pnas.83.4.867. PMC 322971. PMID 3513164.
- ^ a b Ejim LJ, Blanchard JE, Koteva KP, Sumerfield R, Elowe NH, Chechetto JD, Brown ED, Junop MS, Wright GD (February 2007). "Inhibitors of bacterial cystathionine beta-lyase: leads for new antimicrobial agents and probes of enzyme structure and function". Journal of Medicinal Chemistry. 50 (4): 755–64. doi:10.1021/jm061132r. PMID 17300162.
- ^ Jastrzębowska K, Gabriel I (February 2015). "Inhibitors of amino acids biosynthesis as antifungal agents". Amino Acids. 47 (2): 227–49. doi:10.1007/s00726-014-1873-1. PMC 4302243. PMID 25408465.
