Paleopolyploidy

[1]Paleopolyploidy is the result of genome duplications which occurred at least several million years ago (MYA). Such an event could either double the genome of a single species (autopolyploidy) or combine those of two species (allopolyploidy). Because of functional redundancy, genes are rapidly silenced or lost from the duplicated genomes. Most paleopolyploids, through evolutionary time, have lost their polyploid status through a process called diploidization, and are currently considered diploids, e.g., baker's yeast,[2] Arabidopsis thaliana,[3] and perhaps humans.[4][5][6][7]
Paleopolyploidy is extensively studied in plant lineages. It has been found that almost all flowering plants have undergone at least one round of genome duplication at some point during their evolutionary history. Ancient genome duplications are also found in the early ancestor of vertebrates (which includes the human lineage) near the origin of the bony fishes, and another in the stem lineage of teleost fishes.[8] Evidence suggests that baker's yeast (Saccharomyces cerevisiae), which has a compact genome, experienced polyploidization during its evolutionary history.
The term mesopolyploid is sometimes used for species that have undergone whole genome multiplication events (whole genome duplication, whole genome triplification, etc.) in more recent history, such as within the last 17 million years.[9]
Eukaryotes
[edit]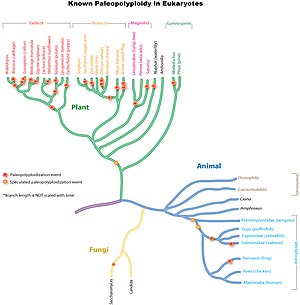
Ancient genome duplications are widespread throughout eukaryotic lineages, particularly in plants. Studies suggest that the common ancestor of Poaceae, the grass family which includes important crop species such as maize, rice, wheat, and sugar cane, shared a whole genome duplication about 70 million years ago.[10] In more ancient monocot lineages one or likely multiple rounds of additional whole genome duplications had occurred, which were however not shared with the ancestral eudicots.[11] Further independent more recent whole genome duplications have occurred in the lineages leading to maize, sugar cane and wheat, but not rice, sorghum or foxtail millet.[citation needed]
A polyploidy event 160 million years ago is theorized to have created the ancestral line that led to all modern flowering plants.[12] That paleopolyploidy event was studied by sequencing the genome of an ancient flowering plant, Amborella trichopoda.[13]
The core eudicots also shared a common whole genome triplication (paleo-hexaploidy), which was estimated to have occurred after monocot-eudicot divergence but before the divergence of rosids and asterids.[14][15][16] Many eudicot species have experienced additional whole genome duplications or triplications. For example, the model plant Arabidopsis thaliana, the first plant to have its entire genome sequenced, has experienced at least two additional rounds of whole genome duplication since the duplication shared by the core eudicots.[3] The most recent event took place before the divergence of the Arabidopsis and Brassica lineages, about 20 million years ago to 45 million years ago. Other examples include the sequenced eudicot genomes of apple, soybean, tomato, cotton, etc.[citation needed]
Compared with plants, paleopolyploidy is much rarer in the animal kingdom. It has been identified mainly in amphibians and bony fishes. Although some studies suggested one or more common genome duplications are shared by all vertebrates (including humans), the evidence is not as strong as in the other cases because the duplications, if they exist, happened so long ago (about 400-500 Ma compared to less than 200 Ma in plants), and the matter is still under debate. The idea that vertebrates share a common whole genome duplication is known as the 2R Hypothesis. Many researchers are interested in the reason why animal lineages, particularly mammals, have had so many fewer whole genome duplications than plant lineages.[citation needed]
A well-supported paleopolyploidy has been found in baker's yeast (Saccharomyces cerevisiae), despite its small, compact genome (~13Mbp), after the divergence from Kluyveromyces lactis and K. marxianus.[17] Through genome streamlining, yeast has lost 90% of the duplicated genome over evolutionary time and is now recognized as a diploid organism.[citation needed]
Detection method
[edit]Duplicated genes can be identified through sequence homology on the DNA or protein level. Paleopolyploidy can be identified as massive gene duplication at one time using a molecular clock. To distinguish between whole-genome duplication and a collection of (more common) single gene duplication events, the following rules are often applied:

- Duplicated genes are located in large duplicated blocks. Single gene duplication is a random process and tends to make duplicated genes scattered throughout the genome.
- Duplicated blocks are non-overlapping because they were created simultaneously. Segmental duplication within the genome can fulfill the first rule; but multiple independent segmental duplications could overlap each other.
In theory, the two duplicated genes should have the same "age"; that is, the divergence of the sequence should be equal between the two genes duplicated by paleopolyploidy (homeologs). Synonymous substitution rate, Ks, is often used as a molecular clock to determine the time of gene duplication. Thus, paleopolyploidy is identified as a "peak" on the duplicate number vs. Ks graph (shown on the right).
However, using Ks plots to identify and document ancient polyploid events can be problematic, as the method fails to identify genome duplications that were followed by massive gene elimination and genome refinement. Other mixed model approaches that combined Ks plots with other methods are being developed to better understand paleopolyploidy.[18]
Duplication events that occurred a long time ago in the history of various evolutionary lineages can be difficult to detect because of subsequent diploidization (such that a polyploid starts to behave cytogenetically as a diploid over time) as mutations and gene translations gradually make one copy of each chromosome unlike its counterpart. This usually results in a low confidence for identifying a very ancient paleopolyploidy.
Evolutionary importance
[edit]Paleopolyploidization events lead to massive cellular changes, including doubling of the genetic material, changes in gene expression and increased cell size. Gene loss during diploidization is not completely random, but heavily selected. Genes from large gene families are duplicated. On the other hand, individual genes are not duplicated.[clarification needed] Overall, paleopolyploidy can have both short-term and long-term evolutionary effects on an organism's fitness in the natural environment.[citation needed]
Enhanced phenotypic evolution
[edit]Whole genome duplication may increase the rates and efficiency by which organisms acquire new biological traits. However, one test of this hypothesis, which compared evolutionary rates in innovation in early teleost fishes (with duplicate genomes) to early holostean fishes (without duplicated genomes) found little difference between the two.[8]
Genome diversity
[edit]Genome doubling provided the organism with redundant alleles that can evolve freely with little selection pressure. The duplicated genes can undergo neofunctionalization or subfunctionalization which could help the organism adapt to the new environment or survive different stress conditions.[citation needed]
Hybrid vigor
[edit]Polyploids often have larger cells and even larger organs. Many important crops, including wheat, maize and cotton, are paleopolyploids which were selected for domestication by ancient peoples.[citation needed]
Speciation
[edit]It has been suggested that many polyploidization events created new species, via a gain of adaptive traits, or by sexual incompatibility with their diploid counterparts. An example would be the recent speciation of allopolyploid Spartina — S. anglica; the polyploid plant is so successful that it is listed as an invasive species in many regions.[19]
Allopolyploidy and autopolyploidy
[edit]There are two major divisions of polyploidy, allopolyploidy and autopolyploidy. Allopolyploids arise as a result of the hybridization of two related species, while autopolyploids arise from the duplication of a species' genome as a result of hybridization of two conspecific parents,[20] or somatic doubling in reproductive tissue of a parent. Allopolyploid species are believed to be much more prevalent in nature,[20] possibly because allopolyploids inherit different genomes, resulting in increased heterozygosity, and therefore higher fitness. These different genomes result in an increased likelihood of large genomic reorganizations,[20][21] which can be either deleterious, or advantageous. Autopolyploidy, however, is generally considered to be a neutral process,[21] though it has been hypothesized that autopolyploidy may serve as a useful mechanism for inducing speciation, and therefore assisting in the ability of an organism to quickly colonize in new habitats without undergoing the time-intensive and costly period of genomic reorganization experienced by allopolyploid species. One common source of autopolyploidy in plants stems from "perfect flowers", which are capable of self-pollination, or "selfing". This, along with errors in meiosis that lead to aneuploidy, can create an environment where autopolyploidy is very likely. This fact can be exploited in a laboratory setting by using colchicine to inhibit chromosome segregation during meiosis, creating synthetic autopolyploid plants.[citation needed]
Following polyploidy events, there are several possible fates for duplicated genes; both copies may be retained as functional genes, change in gene function may occur in one or both copies, gene silencing may mask one or both copies, or complete gene loss may occur.[20][22] Polyploidy events will result in higher levels of heterozygosity, and, over time, can lead to an increase in the total number of functional genes in the genome. As time passes after a genome duplication event, many genes will change function as a result of either change in duplicate gene function for both allo- and autopolyploid species, or there will be changes in gene expression caused by genomic rearrangements induced by genome duplication in allopolyploids. When both copies of a gene are retained, and thus the number of copies doubled, there is a chance that there will be a proportional increase in expression of that gene, resulting in twice as much mRNA transcript being produced. There is also the possibility that transcription of a duplicated gene will be down-regulated, resulting in less than two-fold increase in transcription of that gene, or that the duplication event will yield more than a two-fold increase in transcription.[23] In one species, Glycine dolichocarpa (a close relative of the soybean, Glycine max), it has been observed that following a genome duplication roughly 500,000 years ago, there has been a 1.4 fold increase in transcription, indicating that there has been a proportional decrease in transcription relative to gene copy number following the duplication event.[23]
Vertebrates as paleopolyploid
[edit]The hypothesis of vertebrate paleopolyploidy originated as early as the 1970s, proposed by the biologist Susumu Ohno. He reasoned that the vertebrate genome could not achieve its complexity without large scale whole-genome duplications. The "two rounds of genome duplication" hypothesis (2R hypothesis) came about, and gained in popularity, especially among developmental biologists.[citation needed]
Some researchers have questioned the 2R hypothesis because it predicts that vertebrate genomes should have a 4:1 gene ratio compared with invertebrate genomes, and this is not supported by findings from the 48 vertebrate genome projects available in mid-2011. For example, the human genome consists of ~21,000 protein coding genes according to June, 2011 counts at UCSC and Ensembl genome analysis centers[citation needed] while an average invertebrate genome size is about 15,000 genes. The amphioxus genome sequence provided support for the hypothesis of two rounds of whole genome duplication, followed by loss of duplicate copies of most genes.[24] Additional arguments against 2R were based on the lack of the (AB)(CD) tree topology amongst four members of a gene family in vertebrates. However, if the two genome duplications occurred close together, we would not expect to find this topology.[25] A recent study generated the sea lamprey genetic map, which yielded strong support for the hypothesis that a single whole-genome duplication occurred in the basal vertebrate lineage, preceded and followed by several evolutionarily independent segmental duplications that occurred over chordate evolution.[26]
See also
[edit]References
[edit]- ^ Garsmeur, Olivier; Schnable, James C; Almeida, Ana; Jourda, Cyril; D’Hont, Angélique; Freeling, Michael (February 1, 2014). "Two evolutionarily distinct classes of paleopolyploidy". Oxford Academic. Retrieved 2024-06-28.
- ^ Kellis M, Birren BW, Lander ES (April 2004). "Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae". Nature. 428 (6983): 617–24. Bibcode:2004Natur.428..617K. doi:10.1038/nature02424. PMID 15004568. S2CID 4422074.
- ^ a b Bowers JE, Chapman BA, Rong J, Paterson AH (March 2003). "Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events". Nature. 422 (6930): 433–8. Bibcode:2003Natur.422..433B. doi:10.1038/nature01521. PMID 12660784. S2CID 4423658.
- ^ Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS, et al. (April 2013). "Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution". Nature Genetics. 45 (4): 415–21, 421e1-2. doi:10.1038/ng.2568. PMC 3709584. PMID 23435085.
- ^ Wolfe KH (May 2001). "Yesterday's polyploids and the mystery of diploidization". Nature Reviews. Genetics. 2 (5): 333–41. doi:10.1038/35072009. PMID 11331899. S2CID 20796914.
- ^ Blanc G, Wolfe KH (July 2004). "Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes". The Plant Cell. 16 (7): 1667–78. doi:10.1105/tpc.021345. PMC 514152. PMID 15208399.
- ^ Blanc G, Wolfe KH (July 2004). "Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution". The Plant Cell. 16 (7): 1679–91. doi:10.1105/tpc.021410. PMC 514153. PMID 15208398.
- ^ a b Clarke JT, Lloyd GT, Friedman M (October 2016). "Little evidence for enhanced phenotypic evolution in early teleosts relative to their living fossil sister group". Proceedings of the National Academy of Sciences of the United States of America. 113 (41): 11531–11536. Bibcode:2016PNAS..11311531C. doi:10.1073/pnas.1607237113. PMC 5068283. PMID 27671652.
- ^ Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, et al. (August 2011). "The genome of the mesopolyploid crop species Brassica rapa". Nature Genetics. 43 (10): 1035–9. doi:10.1038/ng.919. PMID 21873998. S2CID 205358099.
- ^ Paterson AH, Bowers JE, Chapman BA (June 2004). "Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics". Proceedings of the National Academy of Sciences of the United States of America. 101 (26): 9903–8. Bibcode:2004PNAS..101.9903P. doi:10.1073/pnas.0307901101. PMC 470771. PMID 15161969.
- ^ Tang H, Bowers JE, Wang X, Paterson AH (January 2010). "Angiosperm genome comparisons reveal early polyploidy in the monocot lineage". Proceedings of the National Academy of Sciences of the United States of America. 107 (1): 472–7. Bibcode:2010PNAS..107..472T. doi:10.1073/pnas.0908007107. PMC 2806719. PMID 19966307.
- ^ Callaway E (December 2013). "Shrub genome reveals secrets of flower power". Nature. doi:10.1038/nature.2013.14426. S2CID 88293665.
- ^ Adams K (December 2013). "Genomics. Genomic clues to the ancestral flowering plant". Science. 342 (6165): 1456–7. Bibcode:2013Sci...342.1456A. doi:10.1126/science.1248709. PMID 24357306. S2CID 206553839.
- ^ Tang H, Wang X, Bowers JE, Ming R, Alam M, Paterson AH (December 2008). "Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps". Genome Research. 18 (12): 1944–54. doi:10.1101/gr.080978.108. PMC 2593578. PMID 18832442.
- ^ Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, et al. (September 2007). "The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla". Nature. 449 (7161): 463–7. Bibcode:2007Natur.449..463J. doi:10.1038/nature06148. hdl:11577/2430527. PMID 17721507.
- ^ Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH (April 2008). "Synteny and collinearity in plant genomes". Science. 320 (5875): 486–8. Bibcode:2008Sci...320..486T. doi:10.1126/science.1153917. PMID 18436778. S2CID 206510918.
- ^ Wong S, Butler G, Wolfe KH (July 2002). "Gene order evolution and paleopolyploidy in hemiascomycete yeasts". Proceedings of the National Academy of Sciences of the United States of America. 99 (14): 9272–7. Bibcode:2002PNAS...99.9272W. doi:10.1073/pnas.142101099. PMC 123130. PMID 12093907.
- ^ Tiley GP, Barker MS, Burleigh JG (November 2018). "Assessing the Performance of Ks Plots for Detecting Ancient Whole Genome Duplications". Genome Biology and Evolution. 10 (11): 2882–2898. doi:10.1093/gbe/evy200. PMC 6225891. PMID 30239709.
- ^ te Beest M, Le Roux JJ, Richardson DM, Brysting AK, Suda J, Kubesová M, Pysek P (January 2012). "The more the better? The role of polyploidy in facilitating plant invasions". Annals of Botany. 109 (1): 19–45. doi:10.1093/aob/mcr277. PMC 3241594. PMID 22040744.
- ^ a b c d Soltis PS, Soltis DE (June 2000). "The role of genetic and genomic attributes in the success of polyploids". Proceedings of the National Academy of Sciences of the United States of America. 97 (13): 7051–7. Bibcode:2000PNAS...97.7051S. doi:10.1073/pnas.97.13.7051. PMC 34383. PMID 10860970.
- ^ a b Parisod C, Holderegger R, Brochmann C (April 2010). "Evolutionary consequences of autopolyploidy". The New Phytologist. 186 (1): 5–17. doi:10.1111/j.1469-8137.2009.03142.x. PMID 20070540.
- ^ Wendel JF (2000). "Genome evolution in polyploids". Plant Molecular Evolution. Vol. 42. pp. 225–249. doi:10.1007/978-94-011-4221-2_12. ISBN 978-94-010-5833-9. PMID 10688139.
{{cite book}}:|journal=ignored (help) - ^ a b Coate JE, Doyle JJ (2010). "Quantifying whole transcriptome size, a prerequisite for understanding transcriptome evolution across species: an example from a plant allopolyploid". Genome Biology and Evolution. 2: 534–46. doi:10.1093/gbe/evq038. PMC 2997557. PMID 20671102.
- ^ Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, et al. (June 2008). "The amphioxus genome and the evolution of the chordate karyotype". Nature. 453 (7198): 1064–71. Bibcode:2008Natur.453.1064P. doi:10.1038/nature06967. PMID 18563158.
- ^ Furlong RF, Holland PW (April 2002). "Were vertebrates octoploid?". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 357 (1420): 531–44. doi:10.1098/rstb.2001.1035. PMC 1692965. PMID 12028790.
- ^ Smith JJ, Keinath MC (August 2015). "The sea lamprey meiotic map improves resolution of ancient vertebrate genome duplications". Genome Research. 25 (8): 1081–90. doi:10.1101/gr.184135.114. PMC 4509993. PMID 26048246.
Further reading
[edit]- Adams KL, Wendel JF (April 2005). "Polyploidy and genome evolution in plants". Current Opinion in Plant Biology. 8 (2): 135–41. doi:10.1016/j.pbi.2005.01.001. PMID 15752992.
- Cui L, Wall PK, Leebens-Mack JH, Lindsay BG, Soltis DE, Doyle JJ, et al. (June 2006). "Widespread genome duplications throughout the history of flowering plants". Genome Research. 16 (6): 738–49. doi:10.1101/gr.4825606. PMC 1479859. PMID 16702410.
- Comai L (November 2005). "The advantages and disadvantages of being polyploid". Nature Reviews. Genetics. 6 (11): 836–46. doi:10.1038/nrg1711. PMID 16304599. S2CID 3329282.
- Eckardt NA (July 2004). "Two genomes are better than one: widespread paleopolyploidy in plants and evolutionary effects". The Plant Cell. 16 (7): 1647–1649. doi:10.1105/tpc.160710. PMC 514149. PMID 15272471.
- Otto SP, Whitton J (2000). "Polyploid incidence and evolution". Annual Review of Genetics. 34 (1): 401–437. CiteSeerX 10.1.1.323.1059. doi:10.1146/annurev.genet.34.1.401. PMID 11092833.
- Makalowski W (May 2001). "Are we polyploids? A brief history of one hypothesis". Genome Research. 11 (5): 667–70. doi:10.1101/gr.188801. PMID 11337465.
- Kellis M, Birren BW, Lander ES (April 2004). "Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae". Nature. 428 (6983): 617–24. Bibcode:2004Natur.428..617K. doi:10.1038/nature02424. PMID 15004568. S2CID 4422074.
