In situ bioremediation
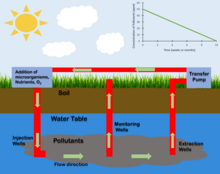
Bioremediation is the process of decontaminating polluted sites through the usage of either endogenous or external microorganism.[1] In situ is a term utilized within a variety of fields meaning "on site" and refers to the location of an event.[2] Within the context of bioremediation, in situ indicates that the location of the bioremediation has occurred at the site of contamination without the translocation of the polluted materials. Bioremediation is used to neutralize pollutants including Hydrocarbons, chlorinated compounds, nitrates, toxic metals and other pollutants through a variety of chemical mechanisms.[1] Microorganism used in the process of bioremediation can either be implanted or cultivated within the site through the application of fertilizers and other nutrients. Common polluted sites targeted by bioremediation are groundwater/aquifers and polluted soils. Aquatic ecosystems affected by oil spills have also shown improvement through the application of bioremediation.[3] The most notable cases being the Deepwater Horizon oil spill in 2010[4] and the Exxon Valdez oil spill in 1989.[5] Two variations of bioremediation exist defined by the location where the process occurs. Ex situ bioremediation occurs at a location separate from the contaminated site and involves the translocation of the contaminated material. In situ occurs within the site of contamination[1] In situ bioremediation can further be categorized by the metabolism occurring, aerobic and anaerobic, and by the level of human involvement.
History
[edit]The Sun Oil pipeline spill in Ambler, Pennsylvania spurred the first commercial usage of in situ bioremediation in 1972 to remove hydrocarbons from contaminated sites.[6] A patent was filed in 1974 by Richard Raymond, Reclamation of Hydrocarbon Contaminated Ground Waters, which provided the basis for the commercialization of in situ bioremediation.[6]
Classifications of In situ Bioremediation
[edit]Accelerated
[edit]Accelerated in situ bioremediation is defined when a specified microorganism is targeted for growth through the application of either nutrients or an electron donor to the contaminated site. Within aerobic metabolism the nutrient added to the soil can be solely Oxygen. Anaerobic in situ bioremediation often requires a variety of electron donors or acceptors such as benzoate and lactate.[7] Besides nutrients, microorganisms can also be introduced directly to the site within accelerated in situ bioremediation.[8] The addition of extraneous microorganisms to a site is termed bioaugmentation and is used when a particular microorganism is effective at degrading the pollutant at the site and is not found either naturally or at a high enough population to be effective.[7] Accelerated in situ bioremediation is utilized when the desired population of microorganisms within a site is not naturally present at a sufficient level to effectively degrade the pollutants. It also is used when the required nutrients within the site are either not at a concentration sufficient to support growth or are unavailable.[7]
Raymond Process
[edit]The Raymond Process is a type of accelerated in situ bioremediation that was developed by Richard Raymond and involves the introduction of nutrients and electron acceptors to a contaminated site.[9] This process is primarily used to treat polluted groundwater. In the Raymond process a loop system is created. Contaminated Groundwater from downstream of the groundwater flow is pumped to the surface and infused with nutrients and an electron donor, often oxygen. This treated water is then pumped back down below the water table upstream of where it was originally taken. This process introduces nutrients and electron donors into the site allowing for the growth of a determined microbial population.[9]
Oxygen Injection
[edit]In contaminated sites where the desired microbial metabolism is aerobic the introduction of oxygen to the site can be used to increase the population of targeted microorganisms.[10] The injection of Oxygen can occur through a variety of processes. Oxygen can be injected into the subsurface through injection wells. It can also be introduced through an injection gallery. The presence of oxygen within a site is often the limiting factor when determining the time frame and efficacy of a proposed in situ bioremediation process.
Ozone Injection
[edit]Ozone injected into the subsurface can also be a means of introducing oxygen into a contaminated site.[10] Despite being a strong oxidizing agent and potentially having a toxic effect on subsurface microbial populations, ozone can be an efficient means of spreading oxygen throughout a site due to its high solubility.[10] Within twenty minutes after being injected into the subsurface, fifty percent of the ozone will have decomposed to Oxygen.[10] Ozone is commonly introduced to the soil in either a dissolved or gaseous state.[10]
Accelerated Anaerobic In situ Bioremediation
[edit]Within accelerated anaerobic in situ bioremediation electron donors and acceptors are introduced into a contaminated site in order to increase the population of anaerobic microorganisms.[9]
Monitored Natural Attenuation (MNA)
[edit]Monitored Natural Attenuation is in situ bioremediation that occurs with little to no human intervention.[11] This process relies on the natural microbial populations sustained within the contaminated sites to over time reduce the contaminants to a desired level.[11] During monitored natural attenuation the site is monitored in order to track the progress of the bioremediation.[11] Monitored natural attenuation is used in sites where the source of contamination is no longer present, often after other more active types of in situ bioremediation have been conducted.[11]
Uses of In situ bioremediation
[edit]Hydrocarbon Degradation
[edit]Naturally occurring within the soil are microbial populations that utilize hydrocarbons as a source of energy and carbon.[9] Upwards to twenty percent of microbial soil populations have the ability to metabolize hydrocarbons.[9] These populations can through either accelerated or natural monitored attenuation be utilized to neutralize within the soil hydrocarbon pollutants. The metabolic mode of hydrocarbon remediation is primarily aerobic.[9] The end products of the remediation for hydrocarbons are Carbon Dioxide and water.[9] Hydrocarbons vary in ease of degradation based on their structure. Long chain aliphatic carbons are the most effectively degraded. Short chained, branched, and quaternary aliphatic hydrocarbons are less effectively degraded.[9] Alkene degradation is dependent on the saturation of the chain with saturated alkenes being more readily degraded.[9] Large numbers of microbes with the ability to metabolize aromatic hydrocarbons are present within the soil. Aromatic hydrocarbons are also susceptible to being degraded through anaerobic metabolism.[9] Hydrocarbon metabolism is an important facet of in situ bioremediation due to the severity of petroleum spills around the world. Polynuclear aromatic carbons susceptibility to degradation is related to the number of aromatic rings within the compound.[9] Compounds with two or three rings are degraded at an effective rate, but compounds possessing four or more rings can be more resilient to bioremediation efforts.[9] Degradation of polynuclear aromatic carbons with less than four rings is accomplished by various aerobic microbes present in the soil. Meanwhile, for larger-molecular-sized compounds, the only metabolic mode that has shown to be effective is cometabolism.[9] The fungus genus Phanerochaete under anaerobic conditions has species with the ability to metabolize some polynuclear aromatic carbons utilizing a peroxidase enzyme.[9][12]
Chlorinated Compounds
[edit]Chlorinated Aliphatic Compounds
[edit]A variety of metabolic modes exist capable of degrading chlorinated aliphatic compounds. Anaerobic reduction, oxidation of the compound, and cometabolism under aerobic conditions are the three main metabolic modes utilized by microorganisms to degrade chlorinated aliphatic compounds.[9] Organisms that can readily metabolize chlorinated aliphatic compounds are not common in the environment.[9] One and two carbons that have little chlorination are the compounds most effectively metabolized by soil microbial populations.[9] The degradation of chlorinated aliphatic compounds is most often performed through cometabolism.[9]

Chlorinated Aromatic Hydrocarbons
[edit]Chlorinated aromatic hydrocarbons are resistant to bioremediation and many microorganisms lack the ability to degrade the compounds. Chlorinated aromatic hydrocarbons are most often degraded through a process of reductive dechlorination under anaerobic conditions.[9] Polychlorinated biphenyls (PCBs) are primarily degraded through cometabolism. There are also some fungi that can degrade these compounds as well. Studies show an increase in PCB degradation when biphenyl is added to the site due to cometabolic effects that the enzymes used to degrade biphenyl have on PCBs.[9]
Benefits
[edit]Due to in situ bioremediation taking place at the site of contamination there is a lessened risk of cross contamination as opposed to ex situ bioremediation where the polluted material is transported to other sites. In situ bioremediation can also have lower costs and a higher rate of decontamination than ex situ bioremediation.
References
[edit]- ^ a b c Rittmann, Bruce E. (1 January 1994). In Situ Bioremediation. Taylor & Francis. ISBN 9780815513483.
- ^ "Charlton T. Lewis, Charles Short, A Latin Dictionary, sĭtus". www.perseus.tufts.edu. Retrieved 4 April 2017.
- ^ Perelo, Louisa Wessels (15 May 2010). "Review: In situ and bioremediation of organic pollutants in aquatic sediments". Journal of Hazardous Materials. 177 (1–3): 81–89. Bibcode:2010JHzM..177...81P. doi:10.1016/j.jhazmat.2009.12.090. PMID 20138425.
- ^ Biello, David. "Slick Solution: How Microbes Will Clean Up the Deepwater Horizon Oil Spill". Scientific American. Retrieved 17 March 2017.
- ^ Atlas, Ronald M.; Hazen, Terry C. (15 August 2011). "Oil Biodegradation and Bioremediation: A Tale of the Two Worst Spills in U.S. History". Environmental Science & Technology. 45 (16): 6709–6715. Bibcode:2011EnST...45.6709A. doi:10.1021/es2013227. ISSN 0013-936X. PMC 3155281. PMID 21699212.
- ^ a b "Lecture 12 Bioremediation" (PDF). Online Course Work Massachusetts Institute of Technology. Retrieved 17 March 2017.
- ^ a b c "In Situ Bioremediation". bioprocess.pnnl.gov. Retrieved 17 March 2017.
- ^ Ellis, David E.; Lutz, Edward J.; Odom, J. Martin; Buchanan, Ronald J.; Bartlett, Craig L.; Lee, Michael D.; Harkness, Mark R.; DeWeerd, Kim A. (1 June 2000). "Bioaugmentation for Accelerated In Situ Anaerobic Bioremediation". Environmental Science & Technology. 34 (11): 2254–2260. Bibcode:2000EnST...34.2254E. doi:10.1021/es990638e. ISSN 0013-936X.
- ^ a b c d e f g h i j k l m n o p q r s t Suthersan, Susan (1999). "IN SITU BIOREMEDIATION" (PDF). University of California Barbara. Retrieved 17 March 2017.
- ^ a b c d e "CLU-IN | Technologies > Remediation > About Remediation Technologies > Bioremediation > Aerobic Bioremediation (Direct)". clu-in.org. Retrieved 17 March 2017.
- ^ a b c d "A Citizen's Guide to Monitored Natural Attenuation" (PDF). Contaminated Sites Clean up Information. September 2012. Retrieved 17 March 2017.
- ^ Syed, Khajamohiddin; Yadav, Jagjit S. (1 November 2012). "P450 monooxygenases (P450ome) of the model white rot fungus Phanerochaete chrysosporium". Critical Reviews in Microbiology. 38 (4): 339–363. doi:10.3109/1040841X.2012.682050. ISSN 1040-841X. PMC 3567848. PMID 22624627.
