Untranslated region
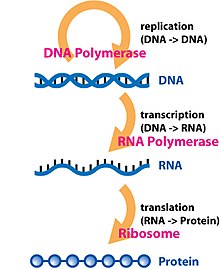

In molecular genetics, an untranslated region (or UTR) refers to either of two sections, one on each side of a coding sequence on a strand of mRNA. If it is found on the 5' side, it is called the 5' UTR (or leader sequence), or if it is found on the 3' side, it is called the 3' UTR (or trailer sequence). mRNA is RNA that carries information from DNA to the ribosome, the site of protein synthesis (translation) within a cell. The mRNA is initially transcribed from the corresponding DNA sequence and then translated into protein. However, several regions of the mRNA are usually not translated into protein, including the 5' and 3' UTRs.
Although they are called untranslated regions, and do not form the protein-coding region of the gene, uORFs located within the 5' UTR can be translated into peptides.[1]
The 5' UTR is upstream from the coding sequence. Within the 5' UTR is a sequence that is recognized by the ribosome which allows the ribosome to bind and initiate translation. The mechanism of translation initiation differs in prokaryotes and eukaryotes. The 3' UTR is found immediately following the translation stop codon. The 3' UTR plays a critical role in translation termination as well as post-transcriptional modification.[2]
These often long sequences were once thought to be useless or junk mRNA that has simply accumulated over evolutionary time. However, it is now known that the untranslated region of mRNA is involved in many regulatory aspects of gene expression in eukaryotic organisms. The importance of these non-coding regions is supported by evolutionary reasoning, as natural selection would have otherwise eliminated this unusable RNA.
It is important to distinguish the 5' and 3' UTRs from other non-protein-coding RNA. Within the coding sequence of pre-mRNA, there can be found sections of RNA that will not be included in the protein product. These sections of RNA are called introns. The RNA that results from RNA splicing is a sequence of exons. The reason why introns are not considered untranslated regions is that the introns are spliced out in the process of RNA splicing. The introns are not included in the mature mRNA molecule that will undergo translation and are thus considered non-protein-coding RNA.
History
[edit]The untranslated regions of mRNA became a subject of study as early as the late 1970s, after the first mRNA molecule was fully sequenced. In 1978, the 5' UTR of the human gamma-globin mRNA was fully sequenced.[3] In 1980, a study was conducted on the 3' UTR of the duplicated human alpha-globin genes.[4]
Evolution
[edit]The untranslated region is seen in prokaryotes and eukaryotes, although the length and composition may vary. In prokaryotes, the 5' UTR is typically between 3 and 10 nucleotides long. In eukaryotes, the 5' UTR can be hundreds to thousands of nucleotides long. This is consistent with the higher complexity of the genomes of eukaryotes compared to prokaryotes. The 3' UTR varies in length as well. The poly-A tail is essential for keeping the mRNA from being degraded. Although there is variation in lengths of both the 5' and 3' UTR, it has been seen that the 5' UTR length is more highly conserved in evolution than the 3' UTR length.[5]
Prokaryotes
[edit]The 5' UTR of prokaryotes consists of the Shine–Dalgarno sequence (5'-AGGAGGU-3').[6] This sequence is found 3-10 base pairs upstream from the initiation codon. The initiation codon is the start site of translation into protein.
Eukaryotes
[edit]The 5' UTR of eukaryotes is more complex than prokaryotes. It contains a Kozak consensus sequence (ACCAUGG).[7] This sequence contains the initiation codon. The initiation codon is the start site of translation into protein.
Links to disease
[edit]The importance of these untranslated regions of mRNA is just beginning to be understood. Various medical studies are being conducted that have found connections between mutations in untranslated regions and increased risk for developing a particular disease, such as cancer. For example, associations between polymorphisms in the HLA-G 3′UTR region and development of colorectal cancer have been discovered.[8] Single Nucleotide Polymorphisms in the 3' UTR of another gene have also been associated with susceptibility to preterm birth.[9] Mutations in the 3' UTR of the APP gene are related to development of cerebral amyloid angiopathy.[10]
Further study
[edit]Through the recent study of untranslated regions, general information has been gathered about the nature and function of these elements. However, there is still much that is unknown about these regions of mRNA. Since the regulation of gene expression is critical in the proper function of cells, this is an area of study that needs to be investigated further. It is important to consider that mutations in 3' untranslated regions have the potential to alter the expression of several genes that may appear unrelated.[11] We are only beginning to understand the links between proper untranslated region function, and disease states of cells.
See also
[edit]- Atlas of UTR Regulatory Activity
- Coding region
- Five prime untranslated region
- History of RNA biology
- MiRNA
- Three prime untranslated region
- Upstream open reading frames (uORFs)
References
[edit]- ^ Vilela, Cristina; McCarthy, John E. G. (2003-08-01). "Regulation of fungal gene expression via short open reading frames in the mRNA 5'untranslated region". Molecular Microbiology. 49 (4): 859–867. doi:10.1046/j.1365-2958.2003.03622.x. ISSN 0950-382X. PMID 12890013.
- ^ Barrett, Lucy W; Fletcher, Sue; Wilton, Steve D (2013). Untranslated Gene Regions and Other Non-Coding Elements. Springer. ISBN 978-3-0348-0679-4.
- ^ Chang, J. C.; Poon, R.; Neumann, K. H.; Kan, Y. W. (1978-10-01). "The nucleotide sequence of the 5' untranslated region of human gamma-globin mRNA". Nucleic Acids Research. 5 (10): 3515–3522. doi:10.1093/nar/5.10.3515. ISSN 0305-1048. PMC 342692. PMID 318162.
- ^ Michelson, A. M.; Orkin, S. H. (1980-11-01). "The 3' untranslated regions of the duplicated human alpha-globin genes are unexpectedly divergent". Cell. 22 (2 Pt 2): 371–377. doi:10.1016/0092-8674(80)90347-5. ISSN 0092-8674. PMID 7448866. S2CID 54238986.
- ^ Lin, Zhenguo; Li, Wen-Hsiung (2012-01-01). "Evolution of 5' untranslated region length and gene expression reprogramming in yeasts". Molecular Biology and Evolution. 29 (1): 81–89. doi:10.1093/molbev/msr143. ISSN 1537-1719. PMC 3245540. PMID 21965341.
- ^ Jin, H; Zhao, Q; Gonzalez; de Valdivia, EI; Ardell, DH; Stenström, M; Isaksson, LA (April 2006). "Influences on gene expression in vivo by a Shine-Dalgarno sequence". Molecular Microbiology. 60 (2): 480–492. doi:10.1111/j.1365-2958.2006.05110.x. PMID 16573696. S2CID 5686240.
- ^ Nakagawa, So; Niimura, Yoshihito; Gojobori, Takashi; Tanaka, Hiroshi; Miura, Kin-ichiro (2008-02-01). "Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes". Nucleic Acids Research. 36 (3): 861–871. doi:10.1093/nar/gkm1102. ISSN 0305-1048. PMC 2241899. PMID 18086709.
- ^ M. Garziera; E. Catamo; S. Crovella; M. Montico; E. Cecchin; S. Lonardi; E. Mini; S. Nobili; L. Romanato; G. Toffoli (2015). "Association of the HLA-G 3′UTR polymorphisms with colorectal cancer in Italy: a first insight". International Journal of Immunogenetics. 43 (1): 32–39. doi:10.1111/iji.12243. PMID 26752414. S2CID 205193023.
- ^ Zhu, Qin; Chen, Ying; Dai, Jianrong; Wang, Benjing; Liu, Minjuan; Wang, Yun; Tao, Jianying; Li, Hong (2015-01-01). "Methylenetetrahydrofolate reductase polymorphisms at 3'-untranslated region are associated with susceptibility to preterm birth". Translational Pediatrics. 4 (1): 57–62. doi:10.3978/j.issn.2224-4336.2015.01.02. ISSN 2224-4344. PMC 4729064. PMID 26835361.
- ^ G. Nicolas; D. Wallon; C. Goupil; A.-C. Richard; C. Pottier; V. Dorval; M. Sarov-Riviere; F. Riant; D. Herve; P. Amouyel; M. Guerchet; B. Ndamba-Bandzouzi; P. Mbelesso; J.-F. Dartigues; J.-C. Lambert; P.-M. Preux; T. Frebourg; D. Campion; D. Hannequin; E. Tournier-Lasserve; S. S. Hebert; A. Rovelet-Lecrux (2016). "Mutation in the 3'untranslated region of APP as a genetic determinant of cerebral amyloid angiopathy". European Journal of Human Genetics. 24 (1): 92–98. doi:10.1038/ejhg.2015.61. PMC 4795229. PMID 25828868.
- ^ Chatterjee, Sangeeta; Pal, Jayanta K. (2009-05-01). "Role of 5′- and 3′-untranslated regions of mRNAs in human diseases". Biology of the Cell. 101 (5): 251–262. doi:10.1042/BC20080104. ISSN 1768-322X. PMID 19275763. S2CID 22689654.
External links
[edit]- Seeley, Rod R.; Stephens, Trent D.; Philip, Tate (2006). "Structure and Function of the Cell - Animation: mRNA Synthesis (Transcription) (Quiz 1)". Anatomy and Physiology (7 ed.). McGraw Hill. ISBN 0072507470.
- Seeley, Rod R.; Stephens, Trent D.; Philip, Tate (2006). "Structure and Function of the Cell - Animation: How Translation Works". Anatomy and Physiology (7 ed.). McGraw Hill. ISBN 0072507470.
- "UTRSite :: Home". utrsite.ba.itb.cnr.it. Archived from the original on Jun 18, 2022.
- Bio125_Molecular Biology (Feb 6, 2015). "Gene Structure". YouTube.
- UTResource
