EF-Tu
| Elongation Factor Thermo Unstable | |||||||||
|---|---|---|---|---|---|---|---|---|---|
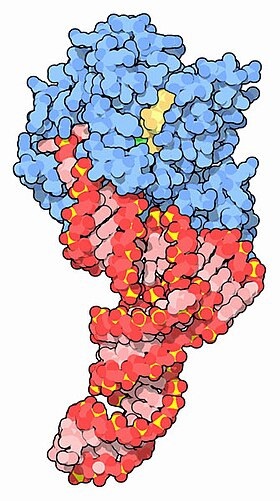 EF-Tu (blue) complexed with tRNA (red) and GTP (yellow) [1] | |||||||||
| Identifiers | |||||||||
| Symbol | EF-Tu | ||||||||
| Pfam | GTP_EFTU | ||||||||
| Pfam clan | CL0023 | ||||||||
| InterPro | IPR004541 | ||||||||
| PROSITE | PDOC00273 | ||||||||
| CATH | 1ETU | ||||||||
| SCOP2 | 1ETU / SCOPe / SUPFAM | ||||||||
| CDD | cd00881 | ||||||||
| |||||||||
| EF-Tu | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | GTP_EFTU_D2 | ||||||||
| Pfam | PF03144 | ||||||||
| InterPro | IPR004161 | ||||||||
| CDD | cd01342 | ||||||||
| |||||||||
| Elongation factor Tu domain 3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | GTP_EFTU_D3 | ||||||||
| Pfam | PF03143 | ||||||||
| InterPro | IPR004160 | ||||||||
| CDD | cd01513 | ||||||||
| |||||||||
EF-Tu (elongation factor thermo unstable) is a prokaryotic elongation factor responsible for catalyzing the binding of an aminoacyl-tRNA (aa-tRNA) to the ribosome. It is a G-protein, and facilitates the selection and binding of an aa-tRNA to the A-site of the ribosome. As a reflection of its crucial role in translation, EF-Tu is one of the most abundant and highly conserved proteins in prokaryotes.[2][3][4] It is found in eukaryotic mitochondria as TUFM.[5]
As a family of elongation factors, EF-Tu also includes its eukaryotic and archaeal homolog, the alpha subunit of eEF-1 (EF-1A).
Background
[edit]Elongation factors are part of the mechanism that synthesizes new proteins through translation in the ribosome. Transfer RNAs (tRNAs) carry the individual amino acids that become integrated into a protein sequence, and have an anticodon for the specific amino acid that they are charged with. Messenger RNA (mRNA) carries the genetic information that encodes the primary structure of a protein, and contains codons that code for each amino acid. The ribosome creates the protein chain by following the mRNA code and integrating the amino acid of an aminoacyl-tRNA (also known as a charged tRNA) to the growing polypeptide chain.[6][7]
There are three sites on the ribosome for tRNA binding. These are the aminoacyl/acceptor site (abbreviated A), the peptidyl site (abbreviated P), and the exit site (abbreviated E). The P-site holds the tRNA connected to the polypeptide chain being synthesized, and the A-site is the binding site for a charged tRNA with an anticodon complementary to the mRNA codon associated with the site. After binding of a charged tRNA to the A-site, a peptide bond is formed between the growing polypeptide chain on the P-site tRNA and the amino acid of the A-site tRNA, and the entire polypeptide is transferred from the P-site tRNA to the A-site tRNA. Then, in a process catalyzed by the prokaryotic elongation factor EF-G (historically known as translocase), the coordinated translocation of the tRNAs and mRNA occurs, with the P-site tRNA moving to the E-site, where it dissociates from the ribosome, and the A-site tRNA moves to take its place in the P-site.[6][7]
Biological functions
[edit]
Protein synthesis
[edit]EF-Tu participates in the polypeptide elongation process of protein synthesis. In prokaryotes, the primary function of EF-Tu is to transport the correct aa-tRNA to the A-site of the ribosome. As a G-protein, it uses GTP to facilitate its function. Outside of the ribosome, EF-Tu complexed with GTP (EF-Tu • GTP) complexes with aa-tRNA to form a stable EF-Tu • GTP • aa-tRNA ternary complex.[8] EF-Tu • GTP binds all correctly-charged aa-tRNAs with approximately identical affinity, except those charged with initiation residues and selenocysteine.[9][10] This can be accomplished because although different amino acid residues have varying side-chain properties, the tRNAs associated with those residues have varying structures to compensate for differences in side-chain binding affinities.[11][12]
The binding of an aa-tRNA to EF-Tu • GTP allows for the ternary complex to be translocated to the A-site of an active ribosome, in which the anticodon of the tRNA binds to the codon of the mRNA. If the correct anticodon binds to the mRNA codon, the ribosome changes configuration and alters the geometry of the GTPase domain of EF-Tu, resulting in the hydrolysis of the GTP associated with the EF-Tu to GDP and Pi. As such, the ribosome functions as a GTPase-activating protein (GAP) for EF-Tu. Upon GTP hydrolysis, the conformation of EF-Tu changes drastically and dissociates from the aa-tRNA and ribosome complex.[4][13] The aa-tRNA then fully enters the A-site, where its amino acid is brought near the P-site's polypeptide and the ribosome catalyzes the covalent transfer of the polypeptide onto the amino acid.[10]
In the cytoplasm, the deactivated EF-Tu • GDP is acted on by the prokaryotic elongation factor EF-Ts, which causes EF-Tu to release its bound GDP. Upon dissociation of EF-Ts, EF-Tu is able to complex with a GTP due to the 5– to 10–fold higher concentration of GTP than GDP in the cytoplasm, resulting in reactivated EF-Tu • GTP, which can then associate with another aa-tRNA.[8][13]
Maintaining translational accuracy
[edit]EF-Tu contributes to translational accuracy in three ways. In translation, a fundamental problem is that near-cognate anticodons have similar binding affinity to a codon as cognate anticodons, such that anticodon-codon binding in the ribosome alone is not sufficient to maintain high translational fidelity. This is addressed by the ribosome not activating the GTPase activity of EF-Tu if the tRNA in the ribosome's A-site does not match the mRNA codon, thus preferentially increasing the likelihood for the incorrect tRNA to leave the ribosome.[14] Additionally, regardless of tRNA matching, EF-Tu also induces a delay after freeing itself from the aa-tRNA, before the aa-tRNA fully enters the A-site (a process called accommodation). This delay period is a second opportunity for incorrectly charged aa-tRNAs to move out of the A-site before the incorrect amino acid is irreversibly added to the polypeptide chain.[15][16] A third mechanism is the less well understood function of EF-Tu to crudely check aa-tRNA associations and reject complexes where the amino acid is not bound to the correct tRNA coding for it.[11]
Other functions
[edit]EF-Tu has been found in large quantities in the cytoskeletons of bacteria, co-localizing underneath the cell membrane with MreB, a cytoskeletal element that maintains cell shape.[17][18] Defects in EF-Tu have been shown to result in defects in bacterial morphology.[19] Additionally, EF-Tu has displayed some chaperone-like characteristics, with some experimental evidence suggesting that it promotes the refolding of a number of denatured proteins in vitro.[20][21] EF-Tu has been found to moonlight on the cell surface of the pathogenic bacteria Staphylococcus aureus, Mycoplasma pneumoniae, and Mycoplasma hyopneumoniae, where EF-Tu is processed and can bind to a range of host molecules.[22] In Bacillus cereus, EF-Tu also moonlights on the surface, where it acts as an environmental sensor and binds to substance P.[23]
Structure
[edit]
EF-Tu is a monomeric protein with molecular weight around 43 kDa in Escherichia coli.[24][25][26] The protein consists of three structural domains: a GTP-binding domain and two oligonucleotide-binding domains, often referred to as domain 2 and domain 3. The N-terminal domain I of EF-Tu is the GTP-binding domain. It consists of a six beta-strand core flanked by six alpha-helices.[8] Domains II and III of EF-Tu, the oligonucleotide-binding domains, both adopt beta-barrel structures.[27][28]
The GTP-binding domain I undergoes a dramatic conformational change upon GTP hydrolysis to GDP, allowing EF-Tu to dissociate from aa-tRNA and leave the ribosome.[29] Reactivation of EF-Tu is achieved by GTP binding in the cytoplasm, which leads to a significant conformational change that reactivates the tRNA-binding site of EF-Tu. In particular, GTP binding to EF-Tu results in a ~90° rotation of domain I relative to domains II and III, exposing the residues of the tRNA-binding active site.[30]
Domain 2 adopts a beta-barrel structure, and is involved in binding to charged tRNA.[31] This domain is structurally related to the C-terminal domain of EF2, to which it displays weak sequence similarity. This domain is also found in other proteins such as translation initiation factor IF-2 and tetracycline-resistance proteins. Domain 3 represents the C-terminal domain, which adopts a beta-barrel structure, and is involved in binding to both charged tRNA and to EF1B (or EF-Ts).[32]
Evolution
[edit]This section may require cleanup to meet Wikipedia's quality standards. The specific problem is: Still terrible, consider just transcluding the section in "further". (December 2023) |
The GTP-binding domain is conserved in both EF-1alpha/EF-Tu and also in EF-2/EF-G and thus seems typical for GTP-dependent proteins which bind non-initiator tRNAs to the ribosome. The GTP-binding translation factor family also includes the eukaryotic peptide chain release factor GTP-binding subunits[33] and prokaryotic peptide chain release factor 3 (RF-3);[34] the prokaryotic GTP-binding protein lepA and its homologue in yeast (GUF1) and Caenorhabditis elegans (ZK1236.1); yeast HBS1;[35] rat Eef1a1 (formerly "statin S1");[36] and the prokaryotic selenocysteine-specific elongation factor selB.[37]
Disease relevance
[edit]Along with the ribosome, EF-Tu is one of the most important targets for antibiotic-mediated inhibition of translation.[8] Antibiotics targeting EF-Tu can be categorized into one of two groups, depending on the mechanism of action, and one of four structural families. The first group includes the antibiotics pulvomycin and GE2270A, and inhibits the formation of the ternary complex.[38] The second group includes the antibiotics kirromycin and enacyloxin, and prevents the release of EF-Tu from the ribosome after GTP hydrolysis.[39][40][41]
See also
[edit]- Prokaryotic elongation factors
- EF-Ts (elongation factor thermo stable)
- EF-G (elongation factor G)
- EF-P (elongation factor P)
- eEF-1
- EFR (EF-Tu receptor)
References
[edit]- ^ PDB Molecule of the Month EF-Tu
- ^ Weijland A, Harmark K, Cool RH, Anborgh PH, Parmeggiani A (March 1992). "Elongation factor Tu: a molecular switch in protein biosynthesis". Molecular Microbiology. 6 (6): 683–8. doi:10.1111/j.1365-2958.1992.tb01516.x. PMID 1573997.
- ^ "TIGR00485: EF-Tu". National Center for Biotechnology Information. March 3, 2017.
- ^ a b Yamamoto H, Qin Y, Achenbach J, Li C, Kijek J, Spahn CM, Nierhaus KH (February 2014). "EF-G and EF4: translocation and back-translocation on the bacterial ribosome". Nature Reviews. Microbiology. 12 (2): 89–100. doi:10.1038/nrmicro3176. PMID 24362468. S2CID 27196901.
- ^ Ling M, Merante F, Chen HS, Duff C, Duncan AM, Robinson BH (Nov 1997). "The human mitochondrial elongation factor tu (EF-Tu) gene: cDNA sequence, genomic localization, genomic structure, and identification of a pseudogene". Gene. 197 (1–2): 325–36. doi:10.1016/S0378-1119(97)00279-5. PMID 9332382.
- ^ a b Laursen BS, Sørensen HP, Mortensen KK, Sperling-Petersen HU (March 2005). "Initiation of protein synthesis in bacteria". Microbiology and Molecular Biology Reviews. 69 (1): 101–23. doi:10.1128/MMBR.69.1.101-123.2005. PMC 1082788. PMID 15755955.
- ^ a b Ramakrishnan V (February 2002). "Ribosome structure and the mechanism of translation". Cell. 108 (4): 557–72. doi:10.1016/s0092-8674(02)00619-0. PMID 11909526. S2CID 2078757.
- ^ a b c d Krab IM, Parmeggiani A (2002-01-01). Mechanisms of EF-Tu, a pioneer GTPase. Vol. 71. pp. 513–51. doi:10.1016/S0079-6603(02)71050-7. ISBN 9780125400718. PMID 12102560.
{{cite book}}:|journal=ignored (help) - ^ "Translation elongation factor EFTu/EF1A, bacterial/organelle (IPR004541)". InterPro.
- ^ a b Diwan, Joyce (2008). "Translation: Protein Synthesis". Rensselaer Polytechnic Institute. Archived from the original on 2017-06-30. Retrieved 2017-03-09.
- ^ a b LaRiviere FJ, Wolfson AD, Uhlenbeck OC (October 2001). "Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation". Science. 294 (5540): 165–8. Bibcode:2001Sci...294..165L. doi:10.1126/science.1064242. PMID 11588263. S2CID 26192336.
- ^ Louie A, Ribeiro NS, Reid BR, Jurnak F (April 1984). "Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP". The Journal of Biological Chemistry. 259 (8): 5010–6. doi:10.1016/S0021-9258(17)42947-4. PMID 6370998.
- ^ a b Clark BF, Nyborg J (February 1997). "The ternary complex of EF-Tu and its role in protein biosynthesis". Current Opinion in Structural Biology. 7 (1): 110–6. doi:10.1016/s0959-440x(97)80014-0. PMID 9032056.
- ^ Nilsson J, Nissen P (June 2005). "Elongation factors on the ribosome". Current Opinion in Structural Biology. 15 (3): 349–54. doi:10.1016/j.sbi.2005.05.004. PMID 15922593.
- ^ Whitford PC, Geggier P, Altman RB, Blanchard SC, Onuchic JN, Sanbonmatsu KY (June 2010). "Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways". RNA. 16 (6): 1196–204. doi:10.1261/rna.2035410. PMC 2874171. PMID 20427512.
- ^ Noel JK, Whitford PC (October 2016). "How EF-Tu can contribute to efficient proofreading of aa-tRNA by the ribosome". Nature Communications. 7: 13314. Bibcode:2016NatCo...713314N. doi:10.1038/ncomms13314. PMC 5095583. PMID 27796304.
- ^ Defeu Soufo HJ, Reimold C, Linne U, Knust T, Gescher J, Graumann PL (February 2010). "Bacterial translation elongation factor EF-Tu interacts and colocalizes with actin-like MreB protein". Proceedings of the National Academy of Sciences of the United States of America. 107 (7): 3163–8. Bibcode:2010PNAS..107.3163D. doi:10.1073/pnas.0911979107. PMC 2840354. PMID 20133608.
- ^ Mayer F (2003-01-01). "Cytoskeletons in prokaryotes". Cell Biology International. 27 (5): 429–38. doi:10.1016/s1065-6995(03)00035-0. PMID 12758091. S2CID 40897586.
- ^ Mayer F (2006-01-01). "Cytoskeletal elements in bacteria Mycoplasma pneumoniae, Thermoanaerobacterium sp., and Escherichia coli as revealed by electron microscopy". Journal of Molecular Microbiology and Biotechnology. 11 (3–5): 228–43. doi:10.1159/000094057. PMID 16983198. S2CID 23701662.
- ^ Richarme G (November 1998). "Protein-disulfide isomerase activity of elongation factor EF-Tu". Biochemical and Biophysical Research Communications. 252 (1): 156–61. doi:10.1006/bbrc.1998.9591. PMID 9813162.
- ^ Kudlicki W, Coffman A, Kramer G, Hardesty B (December 1997). "Renaturation of rhodanese by translational elongation factor (EF) Tu. Protein refolding by EF-Tu flexing". The Journal of Biological Chemistry. 272 (51): 32206–10. doi:10.1074/jbc.272.51.32206. PMID 9405422.
- ^ Widjaja, Michael; Harvey, Kate Louise; Hagemann, Lisa; Berry, Iain James; Jarocki, Veronica Maria; Raymond, Benjamin Bernard Armando; Tacchi, Jessica Leigh; Gründel, Anne; Steele, Joel Ricky; Padula, Matthew Paul; Charles, Ian George; Dumke, Roger; Djordjevic, Steven Philip (2017-09-11). "Elongation factor Tu is a multifunctional and processed moonlighting protein". Scientific Reports. 7 (1): 11227. doi:10.1038/s41598-017-10644-z. ISSN 2045-2322. PMC 5593925.
- ^ N’Diaye, Awa R.; Borrel, Valerie; Racine, Pierre-Jean; Clamens, Thomas; Depayras, Segolene; Maillot, Olivier; Schaack, Beatrice; Chevalier, Sylvie; Lesouhaitier, Olivier; Feuilloley, Marc G. J. (2019-02-04). "Mechanism of action of the moonlighting protein EfTu as a Substance P sensor in Bacillus cereus". Scientific Reports. 9 (1): 1304. doi:10.1038/s41598-018-37506-6. ISSN 2045-2322. PMC 6361937.
 This article incorporates text from this source, which is available under the CC BY 4.0 license.
This article incorporates text from this source, which is available under the CC BY 4.0 license.
- ^ Caldas TD, El Yaagoubi A, Kohiyama M, Richarme G (October 1998). "Purification of elongation factors EF-Tu and EF-G from Escherichia coli by covalent chromatography on thiol-sepharose". Protein Expression and Purification. 14 (1): 65–70. doi:10.1006/prep.1998.0922. PMID 9758752.
- ^ Wiborg O, Andersen C, Knudsen CR, Clark BF, Nyborg J (August 1996). "Mapping Escherichia coli elongation factor Tu residues involved in binding of aminoacyl-tRNA". The Journal of Biological Chemistry. 271 (34): 20406–11. doi:10.1074/jbc.271.34.20406. PMID 8702777.
- ^ Wurmbach P, Nierhaus KH (1979-01-01). "Isolation of the protein synthesis elongation factors EF-Tu, EF-Ts, and EF-G from Escherichia coli". Nucleic Acids and Protein Synthesis Part H. Methods in Enzymology. Vol. 60. pp. 593–606. doi:10.1016/s0076-6879(79)60056-3. ISBN 9780121819606. PMID 379535.
- ^ Wang Y, Jiang Y, Meyering-Voss M, Sprinzl M, Sigler PB (August 1997). "Crystal structure of the EF-Tu.EF-Ts complex from Thermus thermophilus". Nature Structural Biology. 4 (8): 650–6. doi:10.1038/nsb0897-650. PMID 9253415. S2CID 10644042.
- ^ Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BF, Nyborg J (December 1995). "Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog". Science. 270 (5241): 1464–72. doi:10.1126/science.270.5241.1464. PMID 7491491. S2CID 24817616.
- ^ Möller W, Schipper A, Amons R (September 1987). "A conserved amino acid sequence around Arg-68 of Artemia elongation factor 1 alpha is involved in the binding of guanine nucleotides and aminoacyl transfer RNAs". Biochimie. 69 (9): 983–9. doi:10.1016/0300-9084(87)90232-x. PMID 3126836.
- ^ Kjeldgaard M, Nissen P, Thirup S, Nyborg J (September 1993). "The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation". Structure. 1 (1): 35–50. doi:10.1016/0969-2126(93)90007-4. PMID 8069622.
- ^ Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BF, Nyborg J (December 1995). "Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog". Science. 270 (5241): 1464–72. doi:10.1126/science.270.5241.1464. PMID 7491491. S2CID 24817616.
- ^ Wang Y, Jiang Y, Meyering-Voss M, Sprinzl M, Sigler PB (August 1997). "Crystal structure of the EF-Tu.EF-Ts complex from Thermus thermophilus". Nat. Struct. Biol. 4 (8): 650–6. doi:10.1038/nsb0897-650. PMID 9253415. S2CID 10644042.
- ^ Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF (September 1995). "The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae". EMBO J. 14 (17): 4365–73. doi:10.1002/j.1460-2075.1995.tb00111.x. PMC 394521. PMID 7556078.
- ^ Grentzmann G, Brechemier-Baey D, Heurgué-Hamard V, Buckingham RH (May 1995). "Function of polypeptide chain release factor RF-3 in Escherichia coli. RF-3 action in termination is predominantly at UGA-containing stop signals". J. Biol. Chem. 270 (18): 10595–600. doi:10.1074/jbc.270.18.10595. PMID 7737996.
- ^ Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA (October 1992). "The translation machinery and 70 kd heat shock protein cooperate in protein synthesis". Cell. 71 (1): 97–105. doi:10.1016/0092-8674(92)90269-I. PMID 1394434. S2CID 7417370.
- ^ Ann DK, Moutsatsos IK, Nakamura T, Lin HH, Mao PL, Lee MJ, Chin S, Liem RK, Wang E (June 1991). "Isolation and characterization of the rat chromosomal gene for a polypeptide (pS1) antigenically related to statin". J. Biol. Chem. 266 (16): 10429–37. doi:10.1016/S0021-9258(18)99243-4. PMID 1709933.
- ^ Forchhammer K, Leinfelder W, Bock A (November 1989). "Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein". Nature. 342 (6248): 453–6. Bibcode:1989Natur.342..453F. doi:10.1038/342453a0. PMID 2531290. S2CID 4251625.
- ^ Selva E, Beretta G, Montanini N, Saddler GS, Gastaldo L, Ferrari P, Lorenzetti R, Landini P, Ripamonti F, Goldstein BP (July 1991). "Antibiotic GE2270 a: a novel inhibitor of bacterial protein synthesis. I. Isolation and characterization". The Journal of Antibiotics. 44 (7): 693–701. doi:10.7164/antibiotics.44.693. PMID 1908853.
- ^ Hogg T, Mesters JR, Hilgenfeld R (February 2002). "Inhibitory mechanisms of antibiotics targeting elongation factor Tu". Current Protein & Peptide Science. 3 (1): 121–31. doi:10.2174/1389203023380855. PMID 12370016.
- ^ Andersen GR, Nissen P, Nyborg J (August 2003). "Elongation factors in protein biosynthesis". Trends in Biochemical Sciences. 28 (8): 434–41. doi:10.1016/S0968-0004(03)00162-2. PMID 12932732.
- ^ Parmeggiani A, Nissen P (August 2006). "Elongation factor Tu-targeted antibiotics: four different structures, two mechanisms of action". FEBS Letters. 580 (19): 4576–81. Bibcode:2006FEBSL.580.4576P. doi:10.1016/j.febslet.2006.07.039. PMID 16876786. S2CID 20811259.
External links
[edit]- Peptide+Elongation+Factor+Tu at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Overview of all the structural information available in the PDB for UniProt: P49410 (Elongation factor Tu, mitochondrial) at the PDBe-KB.
