Evolution
| Part of the Biology series on |
| Evolution |
|---|
 |
| Mechanisms and processes |
| Research and history |
| Evolutionary biology fields |
Evolution is the change in the heritable characteristics of biological populations over successive generations.[1][2] It occurs when evolutionary processes such as natural selection and genetic drift act on genetic variation, resulting in certain characteristics becoming more or less common within a population over successive generations.[3] The process of evolution has given rise to biodiversity at every level of biological organisation.[4][5]
The scientific theory of evolution by natural selection was conceived independently by two British naturalists, Charles Darwin and Alfred Russel Wallace, in the mid-19th century as an explanation for why organisms are adapted to their physical and biological environments. The theory was first set out in detail in Darwin's book On the Origin of Species.[6] Evolution by natural selection is established by observable facts about living organisms: (1) more offspring are often produced than can possibly survive; (2) traits vary among individuals with respect to their morphology, physiology, and behaviour; (3) different traits confer different rates of survival and reproduction (differential fitness); and (4) traits can be passed from generation to generation (heritability of fitness).[7] In successive generations, members of a population are therefore more likely to be replaced by the offspring of parents with favourable characteristics for that environment.
In the early 20th century, competing ideas of evolution were refuted and evolution was combined with Mendelian inheritance and population genetics to give rise to modern evolutionary theory.[8] In this synthesis the basis for heredity is in DNA molecules that pass information from generation to generation. The processes that change DNA in a population include natural selection, genetic drift, mutation, and gene flow.[3]
All life on Earth—including humanity—shares a last universal common ancestor (LUCA),[9][10][11] which lived approximately 3.5–3.8 billion years ago.[12] The fossil record includes a progression from early biogenic graphite[13] to microbial mat fossils[14][15][16] to fossilised multicellular organisms. Existing patterns of biodiversity have been shaped by repeated formations of new species (speciation), changes within species (anagenesis), and loss of species (extinction) throughout the evolutionary history of life on Earth.[17] Morphological and biochemical traits tend to be more similar among species that share a more recent common ancestor, which historically was used to reconstruct phylogenetic trees, although direct comparison of genetic sequences is a more common method today.[18][19]
Evolutionary biologists have continued to study various aspects of evolution by forming and testing hypotheses as well as constructing theories based on evidence from the field or laboratory and on data generated by the methods of mathematical and theoretical biology. Their discoveries have influenced not just the development of biology but also other fields including agriculture, medicine, and computer science.[20]
Heredity

Evolution in organisms occurs through changes in heritable characteristics—the inherited characteristics of an organism. In humans, for example, eye colour is an inherited characteristic and an individual might inherit the "brown-eye trait" from one of their parents.[21] Inherited traits are controlled by genes and the complete set of genes within an organism's genome (genetic material) is called its genotype.[22]
The complete set of observable traits that make up the structure and behaviour of an organism is called its phenotype. Some of these traits come from the interaction of its genotype with the environment while others are neutral.[23] Some observable characteristics are not inherited. For example, suntanned skin comes from the interaction between a person's genotype and sunlight; thus, suntans are not passed on to people's children. The phenotype is the ability of the skin to tan when exposed to sunlight. However, some people tan more easily than others, due to differences in genotypic variation; a striking example are people with the inherited trait of albinism, who do not tan at all and are very sensitive to sunburn.[24]
Heritable characteristics are passed from one generation to the next via DNA, a molecule that encodes genetic information.[22] DNA is a long biopolymer composed of four types of bases. The sequence of bases along a particular DNA molecule specifies the genetic information, in a manner similar to a sequence of letters spelling out a sentence. Before a cell divides, the DNA is copied, so that each of the resulting two cells will inherit the DNA sequence. Portions of a DNA molecule that specify a single functional unit are called genes; different genes have different sequences of bases. Within cells, each long strand of DNA is called a chromosome. The specific location of a DNA sequence within a chromosome is known as a locus. If the DNA sequence at a locus varies between individuals, the different forms of this sequence are called alleles. DNA sequences can change through mutations, producing new alleles. If a mutation occurs within a gene, the new allele may affect the trait that the gene controls, altering the phenotype of the organism.[25] However, while this simple correspondence between an allele and a trait works in some cases, most traits are influenced by multiple genes in a quantitative or epistatic manner.[26][27]
Sources of variation
Evolution can occur if there is genetic variation within a population. Variation comes from mutations in the genome, reshuffling of genes through sexual reproduction and migration between populations (gene flow). Despite the constant introduction of new variation through mutation and gene flow, most of the genome of a species is very similar among all individuals of that species.[28] However, discoveries in the field of evolutionary developmental biology have demonstrated that even relatively small differences in genotype can lead to dramatic differences in phenotype both within and between species.
An individual organism's phenotype results from both its genotype and the influence of the environment it has lived in.[27] The modern evolutionary synthesis defines evolution as the change over time in this genetic variation. The frequency of one particular allele will become more or less prevalent relative to other forms of that gene. Variation disappears when a new allele reaches the point of fixation—when it either disappears from the population or replaces the ancestral allele entirely.[29]
Mutation

Mutations are changes in the DNA sequence of a cell's genome and are the ultimate source of genetic variation in all organisms.[30] When mutations occur, they may alter the product of a gene, or prevent the gene from functioning, or have no effect.
About half of the mutations in the coding regions of protein-coding genes are deleterious — the other half are neutral. A small percentage of the total mutations in this region confer a fitness benefit.[31] Some of the mutations in other parts of the genome are deleterious but the vast majority are neutral. A few are beneficial.
Mutations can involve large sections of a chromosome becoming duplicated (usually by genetic recombination), which can introduce extra copies of a gene into a genome.[32] Extra copies of genes are a major source of the raw material needed for new genes to evolve.[33] This is important because most new genes evolve within gene families from pre-existing genes that share common ancestors.[34] For example, the human eye uses four genes to make structures that sense light: three for colour vision and one for night vision; all four are descended from a single ancestral gene.[35]
New genes can be generated from an ancestral gene when a duplicate copy mutates and acquires a new function. This process is easier once a gene has been duplicated because it increases the redundancy of the system; one gene in the pair can acquire a new function while the other copy continues to perform its original function.[36][37] Other types of mutations can even generate entirely new genes from previously noncoding DNA, a phenomenon termed de novo gene birth.[38][39]
The generation of new genes can also involve small parts of several genes being duplicated, with these fragments then recombining to form new combinations with new functions (exon shuffling).[40][41] When new genes are assembled from shuffling pre-existing parts, domains act as modules with simple independent functions, which can be mixed together to produce new combinations with new and complex functions.[42] For example, polyketide synthases are large enzymes that make antibiotics; they contain up to 100 independent domains that each catalyse one step in the overall process, like a step in an assembly line.[43]
One example of mutation is wild boar piglets. They are camouflage coloured and show a characteristic pattern of dark and light longitudinal stripes. However, mutations in the melanocortin 1 receptor (MC1R) disrupt the pattern. The majority of pig breeds carry MC1R mutations disrupting wild-type colour and different mutations causing dominant black colouring.[44]
Sex and recombination
In asexual organisms, genes are inherited together, or linked, as they cannot mix with genes of other organisms during reproduction. In contrast, the offspring of sexual organisms contain random mixtures of their parents' chromosomes that are produced through independent assortment. In a related process called homologous recombination, sexual organisms exchange DNA between two matching chromosomes.[45] Recombination and reassortment do not alter allele frequencies, but instead change which alleles are associated with each other, producing offspring with new combinations of alleles.[46] Sex usually increases genetic variation and may increase the rate of evolution.[47][48]

The two-fold cost of sex was first described by John Maynard Smith.[49] The first cost is that in sexually dimorphic species only one of the two sexes can bear young. This cost does not apply to hermaphroditic species, like most plants and many invertebrates. The second cost is that any individual who reproduces sexually can only pass on 50% of its genes to any individual offspring, with even less passed on as each new generation passes.[50] Yet sexual reproduction is the more common means of reproduction among eukaryotes and multicellular organisms. The Red Queen hypothesis has been used to explain the significance of sexual reproduction as a means to enable continual evolution and adaptation in response to coevolution with other species in an ever-changing environment.[50][51][52][53] Another hypothesis is that sexual reproduction is primarily an adaptation for promoting accurate recombinational repair of damage in germline DNA, and that increased diversity is a byproduct of this process that may sometimes be adaptively beneficial.[54][55]
Gene flow
Gene flow is the exchange of genes between populations and between species.[56] It can therefore be a source of variation that is new to a population or to a species. Gene flow can be caused by the movement of individuals between separate populations of organisms, as might be caused by the movement of mice between inland and coastal populations, or the movement of pollen between heavy-metal-tolerant and heavy-metal-sensitive populations of grasses.
Gene transfer between species includes the formation of hybrid organisms and horizontal gene transfer. Horizontal gene transfer is the transfer of genetic material from one organism to another organism that is not its offspring; this is most common among bacteria.[57] In medicine, this contributes to the spread of antibiotic resistance, as when one bacteria acquires resistance genes it can rapidly transfer them to other species.[58] Horizontal transfer of genes from bacteria to eukaryotes such as the yeast Saccharomyces cerevisiae and the adzuki bean weevil Callosobruchus chinensis has occurred.[59][60] An example of larger-scale transfers are the eukaryotic bdelloid rotifers, which have received a range of genes from bacteria, fungi and plants.[61] Viruses can also carry DNA between organisms, allowing transfer of genes even across biological domains.[62]
Large-scale gene transfer has also occurred between the ancestors of eukaryotic cells and bacteria, during the acquisition of chloroplasts and mitochondria. It is possible that eukaryotes themselves originated from horizontal gene transfers between bacteria and archaea.[63]
Epigenetics
Some heritable changes cannot be explained by changes to the sequence of nucleotides in the DNA. These phenomena are classed as epigenetic inheritance systems.[64] DNA methylation marking chromatin, self-sustaining metabolic loops, gene silencing by RNA interference and the three-dimensional conformation of proteins (such as prions) are areas where epigenetic inheritance systems have been discovered at the organismic level.[65] Developmental biologists suggest that complex interactions in genetic networks and communication among cells can lead to heritable variations that may underlay some of the mechanics in developmental plasticity and canalisation.[66] Heritability may also occur at even larger scales. For example, ecological inheritance through the process of niche construction is defined by the regular and repeated activities of organisms in their environment. This generates a legacy of effects that modify and feed back into the selection regime of subsequent generations.[67] Other examples of heritability in evolution that are not under the direct control of genes include the inheritance of cultural traits and symbiogenesis.[68][69]
Evolutionary forces

From a neo-Darwinian perspective, evolution occurs when there are changes in the frequencies of alleles within a population of interbreeding organisms,[70] for example, the allele for black colour in a population of moths becoming more common. Mechanisms that can lead to changes in allele frequencies include natural selection, genetic drift, and mutation bias.
Natural selection
Evolution by natural selection is the process by which traits that enhance survival and reproduction become more common in successive generations of a population. It embodies three principles:[7]
- Variation exists within populations of organisms with respect to morphology, physiology and behaviour (phenotypic variation).
- Different traits confer different rates of survival and reproduction (differential fitness).
- These traits can be passed from generation to generation (heritability of fitness).
More offspring are produced than can possibly survive, and these conditions produce competition between organisms for survival and reproduction. Consequently, organisms with traits that give them an advantage over their competitors are more likely to pass on their traits to the next generation than those with traits that do not confer an advantage.[71] This teleonomy is the quality whereby the process of natural selection creates and preserves traits that are seemingly fitted for the functional roles they perform.[72] Consequences of selection include nonrandom mating[73] and genetic hitchhiking.
The central concept of natural selection is the evolutionary fitness of an organism.[74] Fitness is measured by an organism's ability to survive and reproduce, which determines the size of its genetic contribution to the next generation.[74] However, fitness is not the same as the total number of offspring: instead fitness is indicated by the proportion of subsequent generations that carry an organism's genes.[75] For example, if an organism could survive well and reproduce rapidly, but its offspring were all too small and weak to survive, this organism would make little genetic contribution to future generations and would thus have low fitness.[74]
If an allele increases fitness more than the other alleles of that gene, then with each generation this allele has a higher probability of becoming common within the population. These traits are said to be "selected for." Examples of traits that can increase fitness are enhanced survival and increased fecundity. Conversely, the lower fitness caused by having a less beneficial or deleterious allele results in this allele likely becoming rarer—they are "selected against."[76]
Importantly, the fitness of an allele is not a fixed characteristic; if the environment changes, previously neutral or harmful traits may become beneficial and previously beneficial traits become harmful.[25] However, even if the direction of selection does reverse in this way, traits that were lost in the past may not re-evolve in an identical form.[77][78] However, a re-activation of dormant genes, as long as they have not been eliminated from the genome and were only suppressed perhaps for hundreds of generations, can lead to the re-occurrence of traits thought to be lost like hindlegs in dolphins, teeth in chickens, wings in wingless stick insects, tails and additional nipples in humans etc. "Throwbacks" such as these are known as atavisms.[79]

· Graph 1 shows directional selection, in which a single extreme phenotype is favoured.
· Graph 2 depicts stabilizing selection, where the intermediate phenotype is favoured over the extreme traits.
· Graph 3 shows disruptive selection, in which the extreme phenotypes are favoured over the intermediate.
Natural selection within a population for a trait that can vary across a range of values, such as height, can be categorised into three different types. The first is directional selection, which is a shift in the average value of a trait over time—for example, organisms slowly getting taller.[80] Secondly, disruptive selection is selection for extreme trait values and often results in two different values becoming most common, with selection against the average value. This would be when either short or tall organisms had an advantage, but not those of medium height. Finally, in stabilising selection there is selection against extreme trait values on both ends, which causes a decrease in variance around the average value and less diversity.[71][81] This would, for example, cause organisms to eventually have a similar height.
Natural selection most generally makes nature the measure against which individuals and individual traits, are more or less likely to survive. "Nature" in this sense refers to an ecosystem, that is, a system in which organisms interact with every other element, physical as well as biological, in their local environment. Eugene Odum, a founder of ecology, defined an ecosystem as: "Any unit that includes all of the organisms...in a given area interacting with the physical environment so that a flow of energy leads to clearly defined trophic structure, biotic diversity, and material cycles (i.e., exchange of materials between living and nonliving parts) within the system...."[82] Each population within an ecosystem occupies a distinct niche, or position, with distinct relationships to other parts of the system. These relationships involve the life history of the organism, its position in the food chain and its geographic range. This broad understanding of nature enables scientists to delineate specific forces which, together, comprise natural selection.
Natural selection can act at different levels of organisation, such as genes, cells, individual organisms, groups of organisms and species.[83][84][85] Selection can act at multiple levels simultaneously.[86] An example of selection occurring below the level of the individual organism are genes called transposons, which can replicate and spread throughout a genome.[87] Selection at a level above the individual, such as group selection, may allow the evolution of cooperation.[88]
Genetic drift

Genetic drift is the random fluctuation of allele frequencies within a population from one generation to the next.[89] When selective forces are absent or relatively weak, allele frequencies are equally likely to drift upward or downward[clarification needed] in each successive generation because the alleles are subject to sampling error.[90] This drift halts when an allele eventually becomes fixed, either by disappearing from the population or by replacing the other alleles entirely. Genetic drift may therefore eliminate some alleles from a population due to chance alone. Even in the absence of selective forces, genetic drift can cause two separate populations that begin with the same genetic structure to drift apart into two divergent populations with different sets of alleles.[91]
According to the neutral theory of molecular evolution most evolutionary changes are the result of the fixation of neutral mutations by genetic drift.[92] In this model, most genetic changes in a population are thus the result of constant mutation pressure and genetic drift.[93] This form of the neutral theory has been debated since it does not seem to fit some genetic variation seen in nature.[94][95] A better-supported version of this model is the nearly neutral theory, according to which a mutation that would be effectively neutral in a small population is not necessarily neutral in a large population.[71] Other theories propose that genetic drift is dwarfed by other stochastic forces in evolution, such as genetic hitchhiking, also known as genetic draft.[90][96][97] Another concept is constructive neutral evolution (CNE), which explains that complex systems can emerge and spread into a population through neutral transitions due to the principles of excess capacity, presuppression, and ratcheting,[98][99][100] and it has been applied in areas ranging from the origins of the spliceosome to the complex interdependence of microbial communities.[101][102][103]
The time it takes a neutral allele to become fixed by genetic drift depends on population size; fixation is more rapid in smaller populations.[104] The number of individuals in a population is not critical, but instead a measure known as the effective population size.[105] The effective population is usually smaller than the total population since it takes into account factors such as the level of inbreeding and the stage of the lifecycle in which the population is the smallest.[105] The effective population size may not be the same for every gene in the same population.[106]
It is usually difficult to measure the relative importance of selection and neutral processes, including drift.[107] The comparative importance of adaptive and non-adaptive forces in driving evolutionary change is an area of current research.[108]
Mutation bias
Mutation bias is usually conceived as a difference in expected rates for two different kinds of mutation, e.g., transition-transversion bias, GC-AT bias, deletion-insertion bias. This is related to the idea of developmental bias. Haldane[109] and Fisher[110] argued that, because mutation is a weak pressure easily overcome by selection, tendencies of mutation would be ineffectual except under conditions of neutral evolution or extraordinarily high mutation rates. This opposing-pressures argument was long used to dismiss the possibility of internal tendencies in evolution,[111] until the molecular era prompted renewed interest in neutral evolution.
Noboru Sueoka[112] and Ernst Freese[113] proposed that systematic biases in mutation might be responsible for systematic differences in genomic GC composition between species. The identification of a GC-biased E. coli mutator strain in 1967,[114] along with the proposal of the neutral theory, established the plausibility of mutational explanations for molecular patterns, which are now common in the molecular evolution literature.
For instance, mutation biases are frequently invoked in models of codon usage.[115] Such models also include effects of selection, following the mutation-selection-drift model,[116] which allows both for mutation biases and differential selection based on effects on translation. Hypotheses of mutation bias have played an important role in the development of thinking about the evolution of genome composition, including isochores.[117] Different insertion vs. deletion biases in different taxa can lead to the evolution of different genome sizes.[118][119] The hypothesis of Lynch regarding genome size relies on mutational biases toward increase or decrease in genome size.
However, mutational hypotheses for the evolution of composition suffered a reduction in scope when it was discovered that (1) GC-biased gene conversion makes an important contribution to composition in diploid organisms such as mammals[120] and (2) bacterial genomes frequently have AT-biased mutation.[121]
Contemporary thinking about the role of mutation biases reflects a different theory from that of Haldane and Fisher. More recent work[111] showed that the original "pressures" theory assumes that evolution is based on standing variation: when evolution depends on events of mutation that introduce new alleles, mutational and developmental biases in the introduction of variation (arrival biases) can impose biases on evolution without requiring neutral evolution or high mutation rates.[111][122] Several studies report that the mutations implicated in adaptation reflect common mutation biases[123][124][125] though others dispute this interpretation.[126]
Genetic hitchhiking
Recombination allows alleles on the same strand of DNA to become separated. However, the rate of recombination is low (approximately two events per chromosome per generation). As a result, genes close together on a chromosome may not always be shuffled away from each other and genes that are close together tend to be inherited together, a phenomenon known as linkage.[127] This tendency is measured by finding how often two alleles occur together on a single chromosome compared to expectations, which is called their linkage disequilibrium. A set of alleles that is usually inherited in a group is called a haplotype. This can be important when one allele in a particular haplotype is strongly beneficial: natural selection can drive a selective sweep that will also cause the other alleles in the haplotype to become more common in the population; this effect is called genetic hitchhiking or genetic draft.[128] Genetic draft caused by the fact that some neutral genes are genetically linked to others that are under selection can be partially captured by an appropriate effective population size.[96]
Sexual selection

A special case of natural selection is sexual selection, which is selection for any trait that increases mating success by increasing the attractiveness of an organism to potential mates.[130] Traits that evolved through sexual selection are particularly prominent among males of several animal species. Although sexually favoured, traits such as cumbersome antlers, mating calls, large body size and bright colours often attract predation, which compromises the survival of individual males.[131][132] This survival disadvantage is balanced by higher reproductive success in males that show these hard-to-fake, sexually selected traits.[133]
Natural outcomes
Evolution influences every aspect of the form and behaviour of organisms. Most prominent are the specific behavioural and physical adaptations that are the outcome of natural selection. These adaptations increase fitness by aiding activities such as finding food, avoiding predators or attracting mates. Organisms can also respond to selection by cooperating with each other, usually by aiding their relatives or engaging in mutually beneficial symbiosis. In the longer term, evolution produces new species through splitting ancestral populations of organisms into new groups that cannot or will not interbreed. These outcomes of evolution are distinguished based on time scale as macroevolution versus microevolution. Macroevolution refers to evolution that occurs at or above the level of species, in particular speciation and extinction, whereas microevolution refers to smaller evolutionary changes within a species or population, in particular shifts in allele frequency and adaptation.[135] Macroevolution is the outcome of long periods of microevolution.[136] Thus, the distinction between micro- and macroevolution is not a fundamental one—the difference is simply the time involved.[137] However, in macroevolution, the traits of the entire species may be important. For instance, a large amount of variation among individuals allows a species to rapidly adapt to new habitats, lessening the chance of it going extinct, while a wide geographic range increases the chance of speciation, by making it more likely that part of the population will become isolated. In this sense, microevolution and macroevolution might involve selection at different levels—with microevolution acting on genes and organisms, versus macroevolutionary processes such as species selection acting on entire species and affecting their rates of speciation and extinction.[138][139][140]
A common misconception is that evolution has goals, long-term plans, or an innate tendency for "progress", as expressed in beliefs such as orthogenesis and evolutionism; realistically, however, evolution has no long-term goal and does not necessarily produce greater complexity.[141][142][143] Although complex species have evolved, they occur as a side effect of the overall number of organisms increasing, and simple forms of life still remain more common in the biosphere.[144] For example, the overwhelming majority of species are microscopic prokaryotes, which form about half the world's biomass despite their small size[145] and constitute the vast majority of Earth's biodiversity.[146] Simple organisms have therefore been the dominant form of life on Earth throughout its history and continue to be the main form of life up to the present day, with complex life only appearing more diverse because it is more noticeable.[147] Indeed, the evolution of microorganisms is particularly important to evolutionary research since their rapid reproduction allows the study of experimental evolution and the observation of evolution and adaptation in real time.[148][149]
Adaptation
Adaptation is the process that makes organisms better suited to their habitat.[150][151] Also, the term adaptation may refer to a trait that is important for an organism's survival. For example, the adaptation of horses' teeth to the grinding of grass. By using the term adaptation for the evolutionary process and adaptive trait for the product (the bodily part or function), the two senses of the word may be distinguished. Adaptations are produced by natural selection.[152] The following definitions are due to Theodosius Dobzhansky:
- Adaptation is the evolutionary process whereby an organism becomes better able to live in its habitat or habitats.[153]
- Adaptedness is the state of being adapted: the degree to which an organism is able to live and reproduce in a given set of habitats.[154]
- An adaptive trait is an aspect of the developmental pattern of the organism which enables or enhances the probability of that organism surviving and reproducing.[155]
Adaptation may cause either the gain of a new feature, or the loss of an ancestral feature. An example that shows both types of change is bacterial adaptation to antibiotic selection, with genetic changes causing antibiotic resistance by both modifying the target of the drug, or increasing the activity of transporters that pump the drug out of the cell.[156] Other striking examples are the bacteria Escherichia coli evolving the ability to use citric acid as a nutrient in a long-term laboratory experiment,[157] Flavobacterium evolving a novel enzyme that allows these bacteria to grow on the by-products of nylon manufacturing,[158][159] and the soil bacterium Sphingobium evolving an entirely new metabolic pathway that degrades the synthetic pesticide pentachlorophenol.[160][161] An interesting but still controversial idea is that some adaptations might increase the ability of organisms to generate genetic diversity and adapt by natural selection (increasing organisms' evolvability).[162][163][164][165]
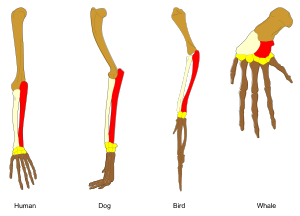
Adaptation occurs through the gradual modification of existing structures. Consequently, structures with similar internal organisation may have different functions in related organisms. This is the result of a single ancestral structure being adapted to function in different ways. The bones within bat wings, for example, are very similar to those in mice feet and primate hands, due to the descent of all these structures from a common mammalian ancestor.[167] However, since all living organisms are related to some extent,[168] even organs that appear to have little or no structural similarity, such as arthropod, squid and vertebrate eyes, or the limbs and wings of arthropods and vertebrates, can depend on a common set of homologous genes that control their assembly and function; this is called deep homology.[169][170]
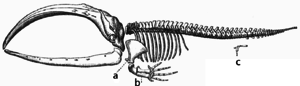
During evolution, some structures may lose their original function and become vestigial structures.[171] Such structures may have little or no function in a current species, yet have a clear function in ancestral species, or other closely related species. Examples include pseudogenes,[172] the non-functional remains of eyes in blind cave-dwelling fish,[173] wings in flightless birds,[174] the presence of hip bones in whales and snakes,[166] and sexual traits in organisms that reproduce via asexual reproduction.[175] Examples of vestigial structures in humans include wisdom teeth,[176] the coccyx,[171] the vermiform appendix,[171] and other behavioural vestiges such as goose bumps[177][178] and primitive reflexes.[179][180][181]
However, many traits that appear to be simple adaptations are in fact exaptations: structures originally adapted for one function, but which coincidentally became somewhat useful for some other function in the process.[182] One example is the African lizard Holaspis guentheri, which developed an extremely flat head for hiding in crevices, as can be seen by looking at its near relatives. However, in this species, the head has become so flattened that it assists in gliding from tree to tree—an exaptation.[182] Within cells, molecular machines such as the bacterial flagella[183] and protein sorting machinery[184] evolved by the recruitment of several pre-existing proteins that previously had different functions.[135] Another example is the recruitment of enzymes from glycolysis and xenobiotic metabolism to serve as structural proteins called crystallins within the lenses of organisms' eyes.[185][186]
An area of current investigation in evolutionary developmental biology is the developmental basis of adaptations and exaptations.[187] This research addresses the origin and evolution of embryonic development and how modifications of development and developmental processes produce novel features.[188] These studies have shown that evolution can alter development to produce new structures, such as embryonic bone structures that develop into the jaw in other animals instead forming part of the middle ear in mammals.[189] It is also possible for structures that have been lost in evolution to reappear due to changes in developmental genes, such as a mutation in chickens causing embryos to grow teeth similar to those of crocodiles.[190] It is now becoming clear that most alterations in the form of organisms are due to changes in a small set of conserved genes.[191]
Coevolution

Interactions between organisms can produce both conflict and cooperation. When the interaction is between pairs of species, such as a pathogen and a host, or a predator and its prey, these species can develop matched sets of adaptations. Here, the evolution of one species causes adaptations in a second species. These changes in the second species then, in turn, cause new adaptations in the first species. This cycle of selection and response is called coevolution.[192] An example is the production of tetrodotoxin in the rough-skinned newt and the evolution of tetrodotoxin resistance in its predator, the common garter snake. In this predator-prey pair, an evolutionary arms race has produced high levels of toxin in the newt and correspondingly high levels of toxin resistance in the snake.[193]
Cooperation
Not all co-evolved interactions between species involve conflict.[194] Many cases of mutually beneficial interactions have evolved. For instance, an extreme cooperation exists between plants and the mycorrhizal fungi that grow on their roots and aid the plant in absorbing nutrients from the soil.[195] This is a reciprocal relationship as the plants provide the fungi with sugars from photosynthesis. Here, the fungi actually grow inside plant cells, allowing them to exchange nutrients with their hosts, while sending signals that suppress the plant immune system.[196]
Coalitions between organisms of the same species have also evolved. An extreme case is the eusociality found in social insects, such as bees, termites and ants, where sterile insects feed and guard the small number of organisms in a colony that are able to reproduce. On an even smaller scale, the somatic cells that make up the body of an animal limit their reproduction so they can maintain a stable organism, which then supports a small number of the animal's germ cells to produce offspring. Here, somatic cells respond to specific signals that instruct them whether to grow, remain as they are, or die. If cells ignore these signals and multiply inappropriately, their uncontrolled growth causes cancer.[197]
Such cooperation within species may have evolved through the process of kin selection, which is where one organism acts to help raise a relative's offspring.[198] This activity is selected for because if the helping individual contains alleles which promote the helping activity, it is likely that its kin will also contain these alleles and thus those alleles will be passed on.[199] Other processes that may promote cooperation include group selection, where cooperation provides benefits to a group of organisms.[200]
Speciation

Speciation is the process where a species diverges into two or more descendant species.[201]
There are multiple ways to define the concept of "species". The choice of definition is dependent on the particularities of the species concerned.[202] For example, some species concepts apply more readily toward sexually reproducing organisms while others lend themselves better toward asexual organisms. Despite the diversity of various species concepts, these various concepts can be placed into one of three broad philosophical approaches: interbreeding, ecological and phylogenetic.[203] The Biological Species Concept (BSC) is a classic example of the interbreeding approach. Defined by evolutionary biologist Ernst Mayr in 1942, the BSC states that "species are groups of actually or potentially interbreeding natural populations, which are reproductively isolated from other such groups."[204] Despite its wide and long-term use, the BSC like other species concepts is not without controversy, for example, because genetic recombination among prokaryotes is not an intrinsic aspect of reproduction;[205] this is called the species problem.[202] Some researchers have attempted a unifying monistic definition of species, while others adopt a pluralistic approach and suggest that there may be different ways to logically interpret the definition of a species.[202][203]
Barriers to reproduction between two diverging sexual populations are required for the populations to become new species. Gene flow may slow this process by spreading the new genetic variants also to the other populations. Depending on how far two species have diverged since their most recent common ancestor, it may still be possible for them to produce offspring, as with horses and donkeys mating to produce mules.[206] Such hybrids are generally infertile. In this case, closely related species may regularly interbreed, but hybrids will be selected against and the species will remain distinct. However, viable hybrids are occasionally formed and these new species can either have properties intermediate between their parent species, or possess a totally new phenotype.[207] The importance of hybridisation in producing new species of animals is unclear, although cases have been seen in many types of animals,[208] with the gray tree frog being a particularly well-studied example.[209]
Speciation has been observed multiple times under both controlled laboratory conditions and in nature.[210] In sexually reproducing organisms, speciation results from reproductive isolation followed by genealogical divergence. There are four primary geographic modes of speciation. The most common in animals is allopatric speciation, which occurs in populations initially isolated geographically, such as by habitat fragmentation or migration. Selection under these conditions can produce very rapid changes in the appearance and behaviour of organisms.[211][212] As selection and drift act independently on populations isolated from the rest of their species, separation may eventually produce organisms that cannot interbreed.[213]
The second mode of speciation is peripatric speciation, which occurs when small populations of organisms become isolated in a new environment. This differs from allopatric speciation in that the isolated populations are numerically much smaller than the parental population. Here, the founder effect causes rapid speciation after an increase in inbreeding increases selection on homozygotes, leading to rapid genetic change.[214]
The third mode is parapatric speciation. This is similar to peripatric speciation in that a small population enters a new habitat, but differs in that there is no physical separation between these two populations. Instead, speciation results from the evolution of mechanisms that reduce gene flow between the two populations.[201] Generally this occurs when there has been a drastic change in the environment within the parental species' habitat. One example is the grass Anthoxanthum odoratum, which can undergo parapatric speciation in response to localised metal pollution from mines.[215] Here, plants evolve that have resistance to high levels of metals in the soil. Selection against interbreeding with the metal-sensitive parental population produced a gradual change in the flowering time of the metal-resistant plants, which eventually produced complete reproductive isolation. Selection against hybrids between the two populations may cause reinforcement, which is the evolution of traits that promote mating within a species, as well as character displacement, which is when two species become more distinct in appearance.[216]
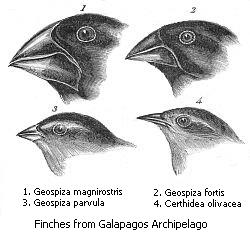
Finally, in sympatric speciation species diverge without geographic isolation or changes in habitat. This form is rare since even a small amount of gene flow may remove genetic differences between parts of a population.[217] Generally, sympatric speciation in animals requires the evolution of both genetic differences and nonrandom mating, to allow reproductive isolation to evolve.[218]
One type of sympatric speciation involves crossbreeding of two related species to produce a new hybrid species. This is not common in animals as animal hybrids are usually sterile. This is because during meiosis the homologous chromosomes from each parent are from different species and cannot successfully pair. However, it is more common in plants because plants often double their number of chromosomes, to form polyploids.[219] This allows the chromosomes from each parental species to form matching pairs during meiosis, since each parent's chromosomes are represented by a pair already.[220] An example of such a speciation event is when the plant species Arabidopsis thaliana and Arabidopsis arenosa crossbred to give the new species Arabidopsis suecica.[221] This happened about 20,000 years ago,[222] and the speciation process has been repeated in the laboratory, which allows the study of the genetic mechanisms involved in this process.[223] Indeed, chromosome doubling within a species may be a common cause of reproductive isolation, as half the doubled chromosomes will be unmatched when breeding with undoubled organisms.[224]
Speciation events are important in the theory of punctuated equilibrium, which accounts for the pattern in the fossil record of short "bursts" of evolution interspersed with relatively long periods of stasis, where species remain relatively unchanged.[225] In this theory, speciation and rapid evolution are linked, with natural selection and genetic drift acting most strongly on organisms undergoing speciation in novel habitats or small populations. As a result, the periods of stasis in the fossil record correspond to the parental population and the organisms undergoing speciation and rapid evolution are found in small populations or geographically restricted habitats and therefore rarely being preserved as fossils.[139]
Extinction

Extinction is the disappearance of an entire species. Extinction is not an unusual event, as species regularly appear through speciation and disappear through extinction.[226] Nearly all animal and plant species that have lived on Earth are now extinct,[227] and extinction appears to be the ultimate fate of all species.[228] These extinctions have happened continuously throughout the history of life, although the rate of extinction spikes in occasional mass extinction events.[229] The Cretaceous–Paleogene extinction event, during which the non-avian dinosaurs became extinct, is the most well-known, but the earlier Permian–Triassic extinction event was even more severe, with approximately 96% of all marine species driven to extinction.[229] The Holocene extinction event is an ongoing mass extinction associated with humanity's expansion across the globe over the past few thousand years. Present-day extinction rates are 100–1000 times greater than the background rate and up to 30% of current species may be extinct by the mid 21st century.[230] Human activities are now the primary cause of the ongoing extinction event;[231][232] global warming may further accelerate it in the future.[233] Despite the estimated extinction of more than 99% of all species that ever lived on Earth,[234][235] about 1 trillion species are estimated to be on Earth currently with only one-thousandth of 1% described.[236]
The role of extinction in evolution is not very well understood and may depend on which type of extinction is considered.[229] The causes of the continuous "low-level" extinction events, which form the majority of extinctions, may be the result of competition between species for limited resources (the competitive exclusion principle).[237] If one species can out-compete another, this could produce species selection, with the fitter species surviving and the other species being driven to extinction.[84] The intermittent mass extinctions are also important, but instead of acting as a selective force, they drastically reduce diversity in a nonspecific manner and promote bursts of rapid evolution and speciation in survivors.[238]
Applications
Concepts and models used in evolutionary biology, such as natural selection, have many applications.[239]
Artificial selection is the intentional selection of traits in a population of organisms. This has been used for thousands of years in the domestication of plants and animals.[240] More recently, such selection has become a vital part of genetic engineering, with selectable markers such as antibiotic resistance genes being used to manipulate DNA. Proteins with valuable properties have evolved by repeated rounds of mutation and selection (for example modified enzymes and new antibodies) in a process called directed evolution.[241]
Understanding the changes that have occurred during an organism's evolution can reveal the genes needed to construct parts of the body, genes which may be involved in human genetic disorders.[242] For example, the Mexican tetra is an albino cavefish that lost its eyesight during evolution. Breeding together different populations of this blind fish produced some offspring with functional eyes, since different mutations had occurred in the isolated populations that had evolved in different caves.[243] This helped identify genes required for vision and pigmentation.[244]
Evolutionary theory has many applications in medicine. Many human diseases are not static phenomena, but capable of evolution. Viruses, bacteria, fungi and cancers evolve to be resistant to host immune defences, as well as to pharmaceutical drugs.[245][246][247] These same problems occur in agriculture with pesticide[248] and herbicide[249] resistance. It is possible that we are facing the end of the effective life of most of available antibiotics[250] and predicting the evolution and evolvability[251] of our pathogens and devising strategies to slow or circumvent it is requiring deeper knowledge of the complex forces driving evolution at the molecular level.[252]
In computer science, simulations of evolution using evolutionary algorithms and artificial life started in the 1960s and were extended with simulation of artificial selection.[253] Artificial evolution became a widely recognised optimisation method as a result of the work of Ingo Rechenberg in the 1960s. He used evolution strategies to solve complex engineering problems.[254] Genetic algorithms in particular became popular through the writing of John Henry Holland.[255] Practical applications also include automatic evolution of computer programmes.[256] Evolutionary algorithms are now used to solve multi-dimensional problems more efficiently than software produced by human designers and also to optimise the design of systems.[257]
Evolutionary history of life
−4500 — – — – −4000 — – — – −3500 — – — – −3000 — – — – −2500 — – — – −2000 — – — – −1500 — – — – −1000 — – — – −500 — – — – 0 — |
| |||||||||||||||||||||||||||||||||||||||||||||
Origin of life
The Earth is about 4.54 billion years old.[258][259][260] The earliest undisputed evidence of life on Earth dates from at least 3.5 billion years ago,[12][261] during the Eoarchean Era after a geological crust started to solidify following the earlier molten Hadean Eon. Microbial mat fossils have been found in 3.48 billion-year-old sandstone in Western Australia.[14][15][16] Other early physical evidence of a biogenic substance is graphite in 3.7 billion-year-old metasedimentary rocks discovered in Western Greenland[13] as well as "remains of biotic life" found in 4.1 billion-year-old rocks in Western Australia.[262][263] Commenting on the Australian findings, Stephen Blair Hedges wrote: "If life arose relatively quickly on Earth, then it could be common in the universe."[262][264] In July 2016, scientists reported identifying a set of 355 genes from the last universal common ancestor (LUCA) of all organisms living on Earth.[265]
More than 99% of all species, amounting to over five billion species,[266] that ever lived on Earth are estimated to be extinct.[234][235] Estimates on the number of Earth's current species range from 10 million to 14 million,[267][268] of which about 1.9 million are estimated to have been named[269] and 1.6 million documented in a central database to date,[270] leaving at least 80% not yet described.
Highly energetic chemistry is thought to have produced a self-replicating molecule around 4 billion years ago, and half a billion years later the last common ancestor of all life existed.[10] The current scientific consensus is that the complex biochemistry that makes up life came from simpler chemical reactions.[271][272] The beginning of life may have included self-replicating molecules such as RNA[273] and the assembly of simple cells.[274]
Common descent
All organisms on Earth are descended from a common ancestor or ancestral gene pool.[168][275] Current species are a stage in the process of evolution, with their diversity the product of a long series of speciation and extinction events.[276] The common descent of organisms was first deduced from four simple facts about organisms: First, they have geographic distributions that cannot be explained by local adaptation. Second, the diversity of life is not a set of completely unique organisms, but organisms that share morphological similarities. Third, vestigial traits with no clear purpose resemble functional ancestral traits. Fourth, organisms can be classified using these similarities into a hierarchy of nested groups, similar to a family tree.[277]

Due to horizontal gene transfer, this "tree of life" may be more complicated than a simple branching tree, since some genes have spread independently between distantly related species.[278][279] To solve this problem and others, some authors prefer to use the "Coral of life" as a metaphor or a mathematical model to illustrate the evolution of life. This view dates back to an idea briefly mentioned by Darwin but later abandoned.[280]
Past species have also left records of their evolutionary history. Fossils, along with the comparative anatomy of present-day organisms, constitute the morphological, or anatomical, record.[281] By comparing the anatomies of both modern and extinct species, palaeontologists can infer the lineages of those species. However, this approach is most successful for organisms that had hard body parts, such as shells, bones or teeth. Further, as prokaryotes such as bacteria and archaea share a limited set of common morphologies, their fossils do not provide information on their ancestry.
More recently, evidence for common descent has come from the study of biochemical similarities between organisms. For example, all living cells use the same basic set of nucleotides and amino acids.[282] The development of molecular genetics has revealed the record of evolution left in organisms' genomes: dating when species diverged through the molecular clock produced by mutations.[283] For example, these DNA sequence comparisons have revealed that humans and chimpanzees share 98% of their genomes and analysing the few areas where they differ helps shed light on when the common ancestor of these species existed.[284]
Evolution of life
Prokaryotes inhabited the Earth from approximately 3–4 billion years ago.[286][287] No obvious changes in morphology or cellular organisation occurred in these organisms over the next few billion years.[288] The eukaryotic cells emerged between 1.6 and 2.7 billion years ago. The next major change in cell structure came when bacteria were engulfed by eukaryotic cells, in a cooperative association called endosymbiosis.[289][290] The engulfed bacteria and the host cell then underwent coevolution, with the bacteria evolving into either mitochondria or hydrogenosomes.[291] Another engulfment of cyanobacterial-like organisms led to the formation of chloroplasts in algae and plants.[292]
The history of life was that of the unicellular eukaryotes, prokaryotes and archaea until about 610 million years ago when multicellular organisms began to appear in the oceans in the Ediacaran period.[286][293] The evolution of multicellularity occurred in multiple independent events, in organisms as diverse as sponges, brown algae, cyanobacteria, slime moulds and myxobacteria.[294] In January 2016, scientists reported that, about 800 million years ago, a minor genetic change in a single molecule called GK-PID may have allowed organisms to go from a single cell organism to one of many cells.[295]
Soon after the emergence of these first multicellular organisms, a remarkable amount of biological diversity appeared over approximately 10 million years, in an event called the Cambrian explosion. Here, the majority of types of modern animals appeared in the fossil record, as well as unique lineages that subsequently became extinct.[296] Various triggers for the Cambrian explosion have been proposed, including the accumulation of oxygen in the atmosphere from photosynthesis.[297]
About 500 million years ago, plants and fungi colonised the land and were soon followed by arthropods and other animals.[298] Insects were particularly successful and even today make up the majority of animal species.[299] Amphibians first appeared around 364 million years ago, followed by early amniotes and birds around 155 million years ago (both from "reptile"-like lineages), mammals around 129 million years ago, Homininae around 10 million years ago and modern humans around 250,000 years ago.[300][301][302] However, despite the evolution of these large animals, smaller organisms similar to the types that evolved early in this process continue to be highly successful and dominate the Earth, with the majority of both biomass and species being prokaryotes.[146]
History of evolutionary thought




Classical antiquity
The proposal that one type of organism could descend from another type goes back to some of the first pre-Socratic Greek philosophers, such as Anaximander and Empedocles.[304] Such proposals survived into Roman times. The poet and philosopher Lucretius followed Empedocles in his masterwork De rerum natura (lit. 'On the Nature of Things').[305][306]
Middle Ages
In contrast to these materialistic views, Aristotelianism had considered all natural things as actualisations of fixed natural possibilities, known as forms.[307][308] This became part of a medieval teleological understanding of nature in which all things have an intended role to play in a divine cosmic order. Variations of this idea became the standard understanding of the Middle Ages and were integrated into Christian learning, but Aristotle did not demand that real types of organisms always correspond one-for-one with exact metaphysical forms and specifically gave examples of how new types of living things could come to be.[309]
A number of Arab Muslim scholars wrote about evolution, most notably Ibn Khaldun, who wrote the book Muqaddimah in 1377 AD, in which he asserted that humans developed from "the world of the monkeys", in a process by which "species become more numerous".[310]
Pre-Darwinian
The "New Science" of the 17th century rejected the Aristotelian approach. It sought to explain natural phenomena in terms of physical laws that were the same for all visible things and that did not require the existence of any fixed natural categories or divine cosmic order. However, this new approach was slow to take root in the biological sciences: the last bastion of the concept of fixed natural types. John Ray applied one of the previously more general terms for fixed natural types, "species", to plant and animal types, but he strictly identified each type of living thing as a species and proposed that each species could be defined by the features that perpetuated themselves generation after generation.[311] The biological classification introduced by Carl Linnaeus in 1735 explicitly recognised the hierarchical nature of species relationships, but still viewed species as fixed according to a divine plan.[312]
Other naturalists of this time speculated on the evolutionary change of species over time according to natural laws. In 1751, Pierre Louis Maupertuis wrote of natural modifications occurring during reproduction and accumulating over many generations to produce new species.[313] Georges-Louis Leclerc, Comte de Buffon, suggested that species could degenerate into different organisms, and Erasmus Darwin proposed that all warm-blooded animals could have descended from a single microorganism (or "filament").[314] The first full-fledged evolutionary scheme was Jean-Baptiste Lamarck's "transmutation" theory of 1809,[315] which envisaged spontaneous generation continually producing simple forms of life that developed greater complexity in parallel lineages with an inherent progressive tendency, and postulated that on a local level, these lineages adapted to the environment by inheriting changes caused by their use or disuse in parents.[316] (The latter process was later called Lamarckism.)[316][317][318] These ideas were condemned by established naturalists as speculation lacking empirical support. In particular, Georges Cuvier insisted that species were unrelated and fixed, their similarities reflecting divine design for functional needs. In the meantime, Ray's ideas of benevolent design had been developed by William Paley into the Natural Theology or Evidences of the Existence and Attributes of the Deity (1802), which proposed complex adaptations as evidence of divine design and which was admired by Charles Darwin.[319][320]
Darwinian revolution
The crucial break from the concept of constant typological classes or types in biology came with the theory of evolution through natural selection, which was formulated by Charles Darwin and Alfred Wallace in terms of variable populations. Darwin used the expression "descent with modification" rather than "evolution".[321] Partly influenced by An Essay on the Principle of Population (1798) by Thomas Robert Malthus, Darwin noted that population growth would lead to a "struggle for existence" in which favourable variations prevailed as others perished. In each generation, many offspring fail to survive to an age of reproduction because of limited resources. This could explain the diversity of plants and animals from a common ancestry through the working of natural laws in the same way for all types of organism.[322][323][324][325] Darwin developed his theory of "natural selection" from 1838 onwards and was writing up his "big book" on the subject when Alfred Russel Wallace sent him a version of virtually the same theory in 1858. Their separate papers were presented together at an 1858 meeting of the Linnean Society of London.[326] At the end of 1859, Darwin's publication of his "abstract" as On the Origin of Species explained natural selection in detail and in a way that led to an increasingly wide acceptance of Darwin's concepts of evolution at the expense of alternative theories. Thomas Henry Huxley applied Darwin's ideas to humans, using paleontology and comparative anatomy to provide strong evidence that humans and apes shared a common ancestry. Some were disturbed by this since it implied that humans did not have a special place in the universe.[327]
Othniel C. Marsh, America’s first paleontologist, was the first to provide solid fossil evidence to support Darwin’s theory of evolution by unearthing the ancestors of the modern horse.[328] In 1877, Marsh delivered a very influential speech before the annual meeting of the American Association for the Advancement of Science, providing a demonstrative argument for evolution. For the first time, Marsh traced the evolution of vertebrates from fish all the way through humans. Sparing no detail, he listed a wealth of fossil examples of past life forms. The significance of this speech was immediately recognized by the scientific community, and it was printed in its entirety in several scientific journals.[329][330]
Pangenesis and heredity
The mechanisms of reproductive heritability and the origin of new traits remained a mystery. Towards this end, Darwin developed his provisional theory of pangenesis.[331] In 1865, Gregor Mendel reported that traits were inherited in a predictable manner through the independent assortment and segregation of elements (later known as genes). Mendel's laws of inheritance eventually supplanted most of Darwin's pangenesis theory.[332] August Weismann made the important distinction between germ cells that give rise to gametes (such as sperm and egg cells) and the somatic cells of the body, demonstrating that heredity passes through the germ line only. Hugo de Vries connected Darwin's pangenesis theory to Weismann's germ/soma cell distinction and proposed that Darwin's pangenes were concentrated in the cell nucleus and when expressed they could move into the cytoplasm to change the cell's structure. De Vries was also one of the researchers who made Mendel's work well known, believing that Mendelian traits corresponded to the transfer of heritable variations along the germline.[333] To explain how new variants originate, de Vries developed a mutation theory that led to a temporary rift between those who accepted Darwinian evolution and biometricians who allied with de Vries.[334][335] In the 1930s, pioneers in the field of population genetics, such as Ronald Fisher, Sewall Wright and J. B. S. Haldane set the foundations of evolution onto a robust statistical philosophy. The false contradiction between Darwin's theory, genetic mutations, and Mendelian inheritance was thus reconciled.[336]
The 'modern synthesis'
In the 1920s and 1930s, the modern synthesis connected natural selection and population genetics, based on Mendelian inheritance, into a unified theory that included random genetic drift, mutation, and gene flow. This new version of evolutionary theory focused on changes in allele frequencies in population. It explained patterns observed across species in populations, through fossil transitions in palaeontology.[336]
Further syntheses
Since then, further syntheses have extended evolution's explanatory power in the light of numerous discoveries, to cover biological phenomena across the whole of the biological hierarchy from genes to populations.[337]
The publication of the structure of DNA by James Watson and Francis Crick with contribution of Rosalind Franklin in 1953 demonstrated a physical mechanism for inheritance.[338] Molecular biology improved understanding of the relationship between genotype and phenotype. Advances were also made in phylogenetic systematics, mapping the transition of traits into a comparative and testable framework through the publication and use of evolutionary trees.[339] In 1973, evolutionary biologist Theodosius Dobzhansky penned that "nothing in biology makes sense except in the light of evolution", because it has brought to light the relations of what first seemed disjointed facts in natural history into a coherent explanatory body of knowledge that describes and predicts many observable facts about life on this planet.[340]
One extension, known as evolutionary developmental biology and informally called "evo-devo", emphasises how changes between generations (evolution) act on patterns of change within individual organisms (development).[237][341] Since the beginning of the 21st century, some biologists have argued for an extended evolutionary synthesis, which would account for the effects of non-genetic inheritance modes, such as epigenetics, parental effects, ecological inheritance and cultural inheritance, and evolvability.[342][343]
Social and cultural responses

In the 19th century, particularly after the publication of On the Origin of Species in 1859, the idea that life had evolved was an active source of academic debate centred on the philosophical, social and religious implications of evolution. Today, the modern evolutionary synthesis is accepted by a vast majority of scientists.[237] However, evolution remains a contentious concept for some theists.[345]
While various religions and denominations have reconciled their beliefs with evolution through concepts such as theistic evolution, there are creationists who believe that evolution is contradicted by the creation myths found in their religions and who raise various objections to evolution.[135][346][347] As had been demonstrated by responses to the publication of Vestiges of the Natural History of Creation in 1844, the most controversial aspect of evolutionary biology is the implication of human evolution that humans share common ancestry with apes and that the mental and moral faculties of humanity have the same types of natural causes as other inherited traits in animals.[348] In some countries, notably the United States, these tensions between science and religion have fuelled the current creation–evolution controversy, a religious conflict focusing on politics and public education.[349] While other scientific fields such as cosmology[350] and Earth science[351] also conflict with literal interpretations of many religious texts, evolutionary biology experiences significantly more opposition from religious literalists.
The teaching of evolution in American secondary school biology classes was uncommon in most of the first half of the 20th century. The Scopes Trial decision of 1925 caused the subject to become very rare in American secondary biology textbooks for a generation, but it was gradually re-introduced later and became legally protected with the 1968 Epperson v. Arkansas decision. Since then, the competing religious belief of creationism was legally disallowed in secondary school curricula in various decisions in the 1970s and 1980s, but it returned in pseudoscientific form as intelligent design (ID), to be excluded once again in the 2005 Kitzmiller v. Dover Area School District case.[352] The debate over Darwin's ideas did not generate significant controversy in China.[353]
See also
- Devolution (biology) – Notion that species can revert to primitive forms
- Chronospecies
References
- ^ Hall & Hallgrímsson 2008, pp. 4–6
- ^ "Evolution Resources". Washington, DC: National Academies of Sciences, Engineering, and Medicine. 2016. Archived from the original on 3 June 2016.
- ^ a b Scott-Phillips, Thomas C.; Laland, Kevin N.; Shuker, David M.; et al. (May 2014). "The Niche Construction Perspective: A Critical Appraisal". Evolution. 68 (5): 1231–1243. doi:10.1111/evo.12332. ISSN 0014-3820. PMC 4261998. PMID 24325256.
Evolutionary processes are generally thought of as processes by which these changes occur. Four such processes are widely recognized: natural selection (in the broad sense, to include sexual selection), genetic drift, mutation, and migration (Fisher 1930; Haldane 1932). The latter two generate variation; the first two sort it.
- ^ Hall & Hallgrímsson 2008, pp. 3–5
- ^ Voet, Voet & Pratt 2016, pp. 1–22, Chapter 1: Introduction to the Chemistry of Life
- ^ Darwin 1859
- ^ a b Lewontin, Richard C. (November 1970). "The Units of Selection" (PDF). Annual Review of Ecology and Systematics. 1: 1–18. Bibcode:1970AnRES...1....1L. doi:10.1146/annurev.es.01.110170.000245. ISSN 0066-4162. JSTOR 2096764. S2CID 84684420. Archived (PDF) from the original on 6 February 2015.
- ^ Futuyma & Kirkpatrick 2017, pp. 3–26, Chapter 1: Evolutionary Biology
- ^ Kampourakis 2014, pp. 127–129
- ^ a b Doolittle, W. Ford (February 2000). "Uprooting the Tree of Life" (PDF). Scientific American. 282 (2): 90–95. Bibcode:2000SciAm.282b..90D. doi:10.1038/scientificamerican0200-90. ISSN 0036-8733. PMID 10710791. Archived from the original (PDF) on 7 September 2006. Retrieved 5 April 2015.
- ^ Glansdorff, Nicolas; Ying Xu; Labedan, Bernard (9 July 2008). "The Last Universal Common Ancestor: emergence, constitution and genetic legacy of an elusive forerunner". Biology Direct. 3: 29. doi:10.1186/1745-6150-3-29. ISSN 1745-6150. PMC 2478661. PMID 18613974.
- ^ a b Schopf, J. William; Kudryavtsev, Anatoliy B.; Czaja, Andrew D.; Tripathi, Abhishek B. (5 October 2007). "Evidence of Archean life: Stromatolites and microfossils". Precambrian Research. 158 (3–4): 141–155. Bibcode:2007PreR..158..141S. doi:10.1016/j.precamres.2007.04.009. ISSN 0301-9268.
- ^ a b Ohtomo, Yoko; Kakegawa, Takeshi; Ishida, Akizumi; et al. (January 2014). "Evidence for biogenic graphite in early Archaean Isua metasedimentary rocks". Nature Geoscience. 7 (1): 25–28. Bibcode:2014NatGe...7...25O. doi:10.1038/ngeo2025. ISSN 1752-0894.
- ^ a b Borenstein, Seth (13 November 2013). "Oldest fossil found: Meet your microbial mom". Excite. Yonkers, New York: Mindspark Interactive Network. Associated Press. Archived from the original on 29 June 2015. Retrieved 31 May 2015.
- ^ a b Pearlman, Jonathan (13 November 2013). "Oldest signs of life on Earth found". The Daily Telegraph. London. Archived from the original on 16 December 2014. Retrieved 15 December 2014.
- ^ a b Noffke, Nora; Christian, Daniel; Wacey, David; Hazen, Robert M. (16 November 2013). "Microbially Induced Sedimentary Structures Recording an Ancient Ecosystem in the ca. 3.48 Billion-Year-Old Dresser Formation, Pilbara, Western Australia". Astrobiology. 13 (12): 1103–1124. Bibcode:2013AsBio..13.1103N. doi:10.1089/ast.2013.1030. ISSN 1531-1074. PMC 3870916. PMID 24205812.
- ^ Futuyma 2004, p. 33
- ^ Panno 2005, pp. xv-16
- ^ NAS 2008, p. 17 Archived 30 June 2015 at the Wayback Machine
- ^ Futuyma, Douglas J., ed. (1999). "Evolution, Science, and Society: Evolutionary Biology and the National Research Agenda" (PDF) (Executive summary). New Brunswick, New Jersey: Office of University Publications, Rutgers, The State University of New Jersey. OCLC 43422991. Archived from the original (PDF) on 31 January 2012. Retrieved 24 November 2014.
- ^ Sturm, Richard A.; Frudakis, Tony N. (August 2004). "Eye colour: portals into pigmentation genes and ancestry". Trends in Genetics. 20 (8): 327–332. doi:10.1016/j.tig.2004.06.010. ISSN 0168-9525. PMID 15262401.
- ^ a b Pearson, Helen (25 May 2006). "Genetics: What is a gene?". Nature. 441 (7092): 398–401. Bibcode:2006Natur.441..398P. doi:10.1038/441398a. ISSN 0028-0836. PMID 16724031. S2CID 4420674.
- ^ Visscher, Peter M.; Hill, William G.; Wray, Naomi R. (April 2008). "Heritability in the genomics era — concepts and misconceptions". Nature Reviews Genetics. 9 (4): 255–266. doi:10.1038/nrg2322. ISSN 1471-0056. PMID 18319743. S2CID 690431.
- ^ Oetting, William S.; Brilliant, Murray H.; King, Richard A. (August 1996). "The clinical spectrum of albinism in humans". Molecular Medicine Today. 2 (8): 330–335. doi:10.1016/1357-4310(96)81798-9. ISSN 1357-4310. PMID 8796918.
- ^ a b Futuyma 2005[page needed]
- ^ Phillips, Patrick C. (November 2008). "Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems". Nature Reviews Genetics. 9 (11): 855–867. doi:10.1038/nrg2452. ISSN 1471-0056. PMC 2689140. PMID 18852697.
- ^ a b Rongling Wu; Min Lin (March 2006). "Functional mapping — how to map and study the genetic architecture of dynamic complex traits". Nature Reviews Genetics. 7 (3): 229–237. doi:10.1038/nrg1804. ISSN 1471-0056. PMID 16485021. S2CID 24301815.
- ^ Butlin, Roger K.; Tregenza, Tom (28 February 1998). "Levels of genetic polymorphism: marker loci versus quantitative traits". Philosophical Transactions of the Royal Society B. 353 (1366): 187–198. doi:10.1098/rstb.1998.0201. ISSN 0962-8436. PMC 1692210. PMID 9533123.
- Butlin, Roger K.; Tregenza, Tom (29 December 2000). "Correction for Butlin and Tregenza, Levels of genetic polymorphism: marker loci versus quantitative traits". Philosophical Transactions of the Royal Society B. 355 (1404): 1865. doi:10.1098/rstb.2000.2000. ISSN 0962-8436.
Some of the values in table 1 on p. 193 were given incorrectly. The errors do not affect the conclusions drawn in the paper. The corrected table is reproduced below.
- Butlin, Roger K.; Tregenza, Tom (29 December 2000). "Correction for Butlin and Tregenza, Levels of genetic polymorphism: marker loci versus quantitative traits". Philosophical Transactions of the Royal Society B. 355 (1404): 1865. doi:10.1098/rstb.2000.2000. ISSN 0962-8436.
- ^ Amos, William; Harwood, John (28 February 1998). "Factors affecting levels of genetic diversity in natural populations". Philosophical Transactions of the Royal Society B. 353 (1366): 177–186. doi:10.1098/rstb.1998.0200. ISSN 0962-8436. PMC 1692205. PMID 9533122.
- ^ Futuyma & Kirkpatrick 2017, pp. 79–102, Chapter 4: Mutation and Variation
- ^ Keightley, PD (2012). "Rates and fitness consequences of new mutations in humans". Genetics. 190 (2): 295–304. doi:10.1534/genetics.111.134668. PMC 3276617. PMID 22345605.
- ^ Hastings, P. J.; Lupski, James R.; Rosenberg, Susan M.; Ira, Grzegorz (August 2009). "Mechanisms of change in gene copy number". Nature Reviews Genetics. 10 (8): 551–564. doi:10.1038/nrg2593. ISSN 1471-0056. PMC 2864001. PMID 19597530.
- ^ Carroll, Grenier & Weatherbee 2005[page needed]
- ^ Harrison, Paul M.; Gerstein, Mark (17 May 2002). "Studying Genomes Through the Aeons: Protein Families, Pseudogenes and Proteome Evolution". Journal of Molecular Biology. 318 (5): 1155–1174. doi:10.1016/S0022-2836(02)00109-2. ISSN 0022-2836. PMID 12083509.
- ^ Bowmaker, James K. (May 1998). "Evolution of colour vision in vertebrates". Eye. 12 (3b): 541–547. doi:10.1038/eye.1998.143. ISSN 0950-222X. PMID 9775215. S2CID 12851209.
- ^ Gregory, T. Ryan; Hebert, Paul D. N. (April 1999). "The Modulation of DNA Content: Proximate Causes and Ultimate Consequences". Genome Research. 9 (4): 317–324. doi:10.1101/gr.9.4.317. ISSN 1088-9051. PMID 10207154. S2CID 16791399. Archived from the original on 23 August 2014. Retrieved 11 December 2014.
- ^ Hurles, Matthew (13 July 2004). "Gene Duplication: The Genomic Trade in Spare Parts". PLOS Biology. 2 (7): e206. doi:10.1371/journal.pbio.0020206. ISSN 1545-7885. PMC 449868. PMID 15252449.
- ^ Liu, Na; Okamura, Katsutomo; Tyler, David M.; et al. (October 2008). "The evolution and functional diversification of animal microRNA genes". Cell Research. 18 (10): 985–996. doi:10.1038/cr.2008.278. ISSN 1001-0602. PMC 2712117. PMID 18711447.
- ^ Siepel, Adam (October 2009). "Darwinian alchemy: Human genes from noncoding DNA". Genome Research. 19 (10): 1693–1695. doi:10.1101/gr.098376.109. ISSN 1088-9051. PMC 2765273. PMID 19797681.
- ^ Orengo, Christine A.; Thornton, Janet M. (July 2005). "Protein families and their evolution—a structural perspective". Annual Review of Biochemistry. 74. Annual Reviews: 867–900. doi:10.1146/annurev.biochem.74.082803.133029. ISSN 0066-4154. PMID 15954844. S2CID 7483470.
- ^ Long, Manyuan; Betrán, Esther; Thornton, Kevin; Wang, Wen (November 2003). "The origin of new genes: glimpses from the young and old". Nature Reviews Genetics. 4 (11): 865–875. doi:10.1038/nrg1204. ISSN 1471-0056. PMID 14634634. S2CID 33999892.
- ^ Wang, Minglei; Caetano-Anollés, Gustavo (14 January 2009). "The Evolutionary Mechanics of Domain Organization in Proteomes and the Rise of Modularity in the Protein World". Structure. 17 (1): 66–78. doi:10.1016/j.str.2008.11.008. ISSN 1357-4310. PMID 19141283.
- ^ Weissman, Kira J.; Müller, Rolf (14 April 2008). "Protein–Protein Interactions in Multienzyme Megasynthetases". ChemBioChem. 9 (6): 826–848. doi:10.1002/cbic.200700751. ISSN 1439-4227. PMID 18357594. S2CID 205552778.
- ^ Andersson, Leif (2020). "Mutations in Domestic Animals Disrupting or Creating Pigmentation Patterns". Frontiers in Ecology and Evolution. 8. doi:10.3389/fevo.2020.00116. ISSN 2296-701X.
- ^ Radding, Charles M. (December 1982). "Homologous Pairing and Strand Exchange in Genetic Recombination". Annual Review of Genetics. 16: 405–437. doi:10.1146/annurev.ge.16.120182.002201. ISSN 0066-4197. PMID 6297377.
- ^ Agrawal, Aneil F. (5 September 2006). "Evolution of Sex: Why Do Organisms Shuffle Their Genotypes?". Current Biology. 16 (17): R696–R704. Bibcode:2006CBio...16.R696A. CiteSeerX 10.1.1.475.9645. doi:10.1016/j.cub.2006.07.063. ISSN 0960-9822. PMID 16950096. S2CID 14739487.
- ^ Peters, Andrew D.; Otto, Sarah P. (June 2003). "Liberating genetic variance through sex". BioEssays. 25 (6): 533–537. doi:10.1002/bies.10291. ISSN 0265-9247. PMID 12766942.
- ^ Goddard, Matthew R.; Godfray, H. Charles J.; Burt, Austin (31 March 2005). "Sex increases the efficacy of natural selection in experimental yeast populations". Nature. 434 (7033): 636–640. Bibcode:2005Natur.434..636G. doi:10.1038/nature03405. ISSN 0028-0836. PMID 15800622. S2CID 4397491.
- ^ Maynard Smith 1978[page needed]
- ^ a b Ridley 2004, p. 314
- ^ Van Valen, Leigh (1973). "A New Evolutionary Law" (PDF). Evolutionary Theory. 1: 1–30. ISSN 0093-4755. Archived from the original (PDF) on 22 December 2014. Retrieved 24 December 2014.
- ^ Hamilton, W. D.; Axelrod, Robert; Tanese, Reiko (1 May 1990). "Sexual reproduction as an adaptation to resist parasites (a review)". PNAS. 87 (9): 3566–3573. Bibcode:1990PNAS...87.3566H. doi:10.1073/pnas.87.9.3566. ISSN 0027-8424. PMC 53943. PMID 2185476.
- ^ Birdsell & Wills 2003, pp. 113–117
- ^ Bernstein H, Byerly HC, Hopf FA, Michod RE. Genetic damage, mutation, and the evolution of sex. Science. 1985 Sep 20;229(4719):1277–81. doi:10.1126/science.3898363. PMID 3898363
- ^ Bernstein H, Hopf FA, Michod RE. The molecular basis of the evolution of sex. Adv Genet. 1987;24:323-70. doi:10.1016/s0065-2660(08)60012-7. PMID 3324702
- ^ Morjan, Carrie L.; Rieseberg, Loren H. (June 2004). "How species evolve collectively: implications of gene flow and selection for the spread of advantageous alleles". Molecular Ecology. 13 (6): 1341–1356. Bibcode:2004MolEc..13.1341M. doi:10.1111/j.1365-294X.2004.02164.x. ISSN 0962-1083. PMC 2600545. PMID 15140081.
- ^ Boucher, Yan; Douady, Christophe J.; Papke, R. Thane; et al. (December 2003). "Lateral gene transfer and the origins of prokaryotic groups". Annual Review of Genetics. 37: 283–328. doi:10.1146/annurev.genet.37.050503.084247. ISSN 0066-4197. PMID 14616063.
- ^ Walsh, Timothy R. (October 2006). "Combinatorial genetic evolution of multiresistance". Current Opinion in Microbiology. 9 (5): 476–482. doi:10.1016/j.mib.2006.08.009. ISSN 1369-5274. PMID 16942901.
- ^ Kondo, Natsuko; Nikoh, Naruo; Ijichi, Nobuyuki; et al. (29 October 2002). "Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect". PNAS. 99 (22): 14280–14285. Bibcode:2002PNAS...9914280K. doi:10.1073/pnas.222228199. ISSN 0027-8424. PMC 137875. PMID 12386340.
- ^ Sprague, George F. Jr. (December 1991). "Genetic exchange between kingdoms". Current Opinion in Genetics & Development. 1 (4): 530–533. doi:10.1016/S0959-437X(05)80203-5. ISSN 0959-437X. PMID 1822285.
- ^ Gladyshev, Eugene A.; Meselson, Matthew; Arkhipova, Irina R. (30 May 2008). "Massive Horizontal Gene Transfer in Bdelloid Rotifers". Science. 320 (5880): 1210–1213. Bibcode:2008Sci...320.1210G. doi:10.1126/science.1156407. ISSN 0036-8075. PMID 18511688. S2CID 11862013. Archived from the original on 30 July 2022. Retrieved 30 July 2022.
- ^ Baldo, Angela M.; McClure, Marcella A. (September 1999). "Evolution and Horizontal Transfer of dUTPase-Encoding Genes in Viruses and Their Hosts". Journal of Virology. 73 (9): 7710–7721. doi:10.1128/JVI.73.9.7710-7721.1999. ISSN 0022-538X. PMC 104298. PMID 10438861.
- ^ Rivera, Maria C.; Lake, James A. (9 September 2004). "The ring of life provides evidence for a genome fusion origin of eukaryotes". Nature. 431 (7005): 152–155. Bibcode:2004Natur.431..152R. doi:10.1038/nature02848. ISSN 0028-0836. PMID 15356622. S2CID 4349149.
- ^ Jablonka, Eva; Raz, Gal (June 2009). "Transgenerational Epigenetic Inheritance: Prevalence, Mechanisms, and Implications for the Study of Heredity and Evolution" (PDF). The Quarterly Review of Biology. 84 (2): 131–176. CiteSeerX 10.1.1.617.6333. doi:10.1086/598822. ISSN 0033-5770. PMID 19606595. S2CID 7233550. Archived (PDF) from the original on 15 July 2011. Retrieved 30 July 2022.
- ^ Bossdorf, Oliver; Arcuri, Davide; Richards, Christina L.; Pigliucci, Massimo (May 2010). "Experimental alteration of DNA methylation affects the phenotypic plasticity of ecologically relevant traits in Arabidopsis thaliana" (PDF). Evolutionary Ecology. 24 (3): 541–553. Bibcode:2010EvEco..24..541B. doi:10.1007/s10682-010-9372-7. ISSN 0269-7653. S2CID 15763479. Archived (PDF) from the original on 5 June 2022. Retrieved 30 July 2022.
- ^ Jablonka, Eva; Lamb, Marion J. (December 2002). "The Changing Concept of Epigenetics". Annals of the New York Academy of Sciences. 981 (1): 82–96. Bibcode:2002NYASA.981...82J. doi:10.1111/j.1749-6632.2002.tb04913.x. ISSN 0077-8923. PMID 12547675. S2CID 12561900.
- ^ Laland, Kevin N.; Sterelny, Kim (September 2006). "Perspective: Seven Reasons (Not) to Neglect Niche Construction". Evolution. 60 (9): 1751–1762. doi:10.1111/j.0014-3820.2006.tb00520.x. ISSN 0014-3820. PMID 17089961. S2CID 22997236.
- ^ Chapman, Michael J.; Margulis, Lynn (December 1998). "Morphogenesis by symbiogenesis" (PDF). International Microbiology. 1 (4): 319–326. ISSN 1139-6709. PMID 10943381. Archived from the original (PDF) on 23 August 2014. Retrieved 9 December 2014.
- ^ Wilson, David Sloan; Wilson, Edward O. (December 2007). "Rethinking the Theoretical Foundation of Sociobiology" (PDF). The Quarterly Review of Biology. 82 (4): 327–348. doi:10.1086/522809. ISSN 0033-5770. PMID 18217526. S2CID 37774648. Archived from the original (PDF) on 11 May 2011.
- ^ Ewens 2004[page needed]
- ^ a b c Hurst, Laurence D. (February 2009). "Fundamental concepts in genetics: genetics and the understanding of selection". Nature Reviews Genetics. 10 (2): 83–93. doi:10.1038/nrg2506. PMID 19119264. S2CID 1670587.
- ^ Darwin 1859, Chapter XIV
- ^ Otto, Sarah P.; Servedio, Maria R.; Nuismer, Scott L. (August 2008). "Frequency-Dependent Selection and the Evolution of Assortative Mating". Genetics. 179 (4): 2091–2112. doi:10.1534/genetics.107.084418. PMC 2516082. PMID 18660541.
- ^ a b c Orr, H. Allen (August 2009). "Fitness and its role in evolutionary genetics". Nature Reviews Genetics. 10 (8): 531–539. doi:10.1038/nrg2603. ISSN 1471-0056. PMC 2753274. PMID 19546856.
- ^ Haldane, J. B. S. (14 March 1959). "The Theory of Natural Selection To-Day". Nature. 183 (4663): 710–713. Bibcode:1959Natur.183..710H. doi:10.1038/183710a0. PMID 13644170. S2CID 4185793.
- ^ Lande, Russell; Arnold, Stevan J. (November 1983). "The Measurement of Selection on Correlated Characters". Evolution. 37 (6): 1210–1226. doi:10.1111/j.1558-5646.1983.tb00236.x. ISSN 0014-3820. JSTOR 2408842. PMID 28556011. S2CID 36544045.
- ^ Goldberg, Emma E.; Igić, Boris (November 2008). "On phylogenetic tests of irreversible evolution". Evolution. 62 (11): 2727–2741. doi:10.1111/j.1558-5646.2008.00505.x. ISSN 0014-3820. PMID 18764918. S2CID 30703407.
- ^ Collin, Rachel; Miglietta, Maria Pia (November 2008). "Reversing opinions on Dollo's Law". Trends in Ecology & Evolution. 23 (11): 602–609. Bibcode:2008TEcoE..23..602C. doi:10.1016/j.tree.2008.06.013. PMID 18814933.
- ^ Tomić, Nenad; Meyer-Rochow, Victor Benno (2011). "Atavisms: Medical, Genetic, and Evolutionary Implications". Perspectives in Biology and Medicine. 54 (3): 332–353. doi:10.1353/pbm.2011.0034. PMID 21857125. S2CID 40851098.
- ^ Hoekstra, Hopi E.; Hoekstra, Jonathan M.; Berrigan, David; et al. (31 July 2001). "Strength and tempo of directional selection in the wild". PNAS. 98 (16): 9157–9160. Bibcode:2001PNAS...98.9157H. doi:10.1073/pnas.161281098. PMC 55389. PMID 11470913.
- ^ Felsenstein, Joseph (November 1979). "Excursions along the Interface between Disruptive and Stabilizing Selection". Genetics. 93 (3): 773–795. doi:10.1093/genetics/93.3.773. PMC 1214112. PMID 17248980.
- ^ Odum 1971, p. 8
- ^ Okasha 2006
- ^ a b Gould, Stephen Jay (28 February 1998). "Gulliver's further travels: the necessity and difficulty of a hierarchical theory of selection". Philosophical Transactions of the Royal Society B. 353 (1366): 307–314. doi:10.1098/rstb.1998.0211. ISSN 0962-8436. PMC 1692213. PMID 9533127.
- ^ Mayr, Ernst (18 March 1997). "The objects of selection". PNAS. 94 (6): 2091–2094. Bibcode:1997PNAS...94.2091M. doi:10.1073/pnas.94.6.2091. ISSN 0027-8424. PMC 33654. PMID 9122151.
- ^ Maynard Smith 1998, pp. 203–211, discussion 211–217
- ^ Hickey, Donal A. (1992). "Evolutionary dynamics of transposable elements in prokaryotes and eukaryotes". Genetica. 86 (1–3): 269–274. doi:10.1007/BF00133725. ISSN 0016-6707. PMID 1334911. S2CID 6583945.
- ^ Gould, Stephen Jay; Lloyd, Elisabeth A. (12 October 1999). "Individuality and adaptation across levels of selection: how shall we name and generalise the unit of Darwinism?". PNAS. 96 (21): 11904–11909. Bibcode:1999PNAS...9611904G. doi:10.1073/pnas.96.21.11904. ISSN 0027-8424. PMC 18385. PMID 10518549.
- ^ Futuyma & Kirkpatrick 2017, pp. 55–66, Chapter 3: Natural Selection and Adaptation
- ^ a b Masel, Joanna (25 October 2011). "Genetic drift". Current Biology. 21 (20): R837–R838. Bibcode:2011CBio...21.R837M. doi:10.1016/j.cub.2011.08.007. ISSN 0960-9822. PMID 22032182. S2CID 17619958.
- ^ Lande, Russell (1989). "Fisherian and Wrightian theories of speciation". Genome. 31 (1): 221–227. doi:10.1139/g89-037. ISSN 0831-2796. PMID 2687093.
- ^ Kimura, Motoo (1991). "The neutral theory of molecular evolution: a review of recent evidence". Japanese Journal of Human Genetics. 66 (4): 367–386. doi:10.1266/jjg.66.367. PMID 1954033. Archived from the original on 5 June 2022.
- ^ Kimura, Motoo (1989). "The neutral theory of molecular evolution and the world view of the neutralists". Genome. 31 (1): 24–31. doi:10.1139/g89-009. ISSN 0831-2796. PMID 2687096.
- ^ Kreitman, Martin (August 1996). "The neutral theory is dead. Long live the neutral theory". BioEssays. 18 (8): 678–683, discussion 683. doi:10.1002/bies.950180812. ISSN 0265-9247. PMID 8760341.
- ^ Leigh, E. G. Jr. (November 2007). "Neutral theory: a historical perspective". Journal of Evolutionary Biology. 20 (6): 2075–2091. doi:10.1111/j.1420-9101.2007.01410.x. ISSN 1010-061X. PMID 17956380. S2CID 2081042.
- ^ a b Gillespie, John H. (November 2001). "Is the population size of a species relevant to its evolution?". Evolution. 55 (11): 2161–2169. doi:10.1111/j.0014-3820.2001.tb00732.x. ISSN 0014-3820. PMID 11794777. S2CID 221735887.
- ^ Neher, Richard A.; Shraiman, Boris I. (August 2011). "Genetic Draft and Quasi-Neutrality in Large Facultatively Sexual Populations". Genetics. 188 (4): 975–996. arXiv:1108.1635. Bibcode:2011arXiv1108.1635N. doi:10.1534/genetics.111.128876. PMC 3176096. PMID 21625002.
- ^ Stoltzfus, Arlin (1999). "On the Possibility of Constructive Neutral Evolution". Journal of Molecular Evolution. 49 (2): 169–181. Bibcode:1999JMolE..49..169S. doi:10.1007/PL00006540. PMID 10441669. S2CID 1743092. Archived from the original on 30 July 2022. Retrieved 30 July 2022.
- ^ Stoltzfus, Arlin (13 October 2012). "Constructive neutral evolution: exploring evolutionary theory's curious disconnect". Biology Direct. 7 (1): 35. doi:10.1186/1745-6150-7-35. PMC 3534586. PMID 23062217.
- ^ Muñoz-Gómez, Sergio A.; Bilolikar, Gaurav; Wideman, Jeremy G.; et al. (1 April 2021). "Constructive Neutral Evolution 20 Years Later". Journal of Molecular Evolution. 89 (3): 172–182. Bibcode:2021JMolE..89..172M. doi:10.1007/s00239-021-09996-y. PMC 7982386. PMID 33604782.
- ^ Lukeš, Julius; Archibald, John M.; Keeling, Patrick J.; et al. (2011). "How a neutral evolutionary ratchet can build cellular complexity". IUBMB Life. 63 (7): 528–537. doi:10.1002/iub.489. PMID 21698757. S2CID 7306575.
- ^ Vosseberg, Julian; Snel, Berend (1 December 2017). "Domestication of self-splicing introns during eukaryogenesis: the rise of the complex spliceosomal machinery". Biology Direct. 12 (1): 30. doi:10.1186/s13062-017-0201-6. PMC 5709842. PMID 29191215.
- ^ Brunet, T. D. P.; Doolittle, W. Ford (19 March 2018). "The generality of Constructive Neutral Evolution". Biology & Philosophy. 33 (1): 2. doi:10.1007/s10539-018-9614-6. S2CID 90290787.
- ^ Otto, Sarah P.; Whitlock, Michael C. (June 1997). "The Probability of Fixation in Populations of Changing Size" (PDF). Genetics. 146 (2): 723–733. doi:10.1093/genetics/146.2.723. PMC 1208011. PMID 9178020. Archived (PDF) from the original on 19 March 2015. Retrieved 18 December 2014.
- ^ a b Charlesworth, Brian (March 2009). "Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation". Nature Reviews Genetics. 10 (3): 195–205. doi:10.1038/nrg2526. PMID 19204717. S2CID 205484393.
- ^ Cutter, Asher D.; Choi, Jae Young (August 2010). "Natural selection shapes nucleotide polymorphism across the genome of the nematode Caenorhabditis briggsae". Genome Research. 20 (8): 1103–1111. doi:10.1101/gr.104331.109. PMC 2909573. PMID 20508143.
- ^ Mitchell-Olds, Thomas; Willis, John H.; Goldstein, David B. (November 2007). "Which evolutionary processes influence natural genetic variation for phenotypic traits?". Nature Reviews Genetics. 8 (11): 845–856. doi:10.1038/nrg2207. ISSN 1471-0056. PMID 17943192. S2CID 14914998.
- ^ Nei, Masatoshi (December 2005). "Selectionism and Neutralism in Molecular Evolution". Molecular Biology and Evolution. 22 (12): 2318–2342. doi:10.1093/molbev/msi242. ISSN 0737-4038. PMC 1513187. PMID 16120807.
- Nei, Masatoshi (May 2006). "Selectionism and Neutralism in Molecular Evolution". Molecular Biology and Evolution (Erratum). 23 (5): 2318–42. doi:10.1093/molbev/msk009. ISSN 0737-4038. PMC 1513187. PMID 16120807.
- ^ Haldane, J.B.S. (July 1927). "A Mathematical Theory of Natural and Artificial Selection, Part V: Selection and Mutation". Proceedings of the Cambridge Philosophical Society. 26 (7): 838–844. Bibcode:1927PCPS...23..838H. doi:10.1017/S0305004100015644. S2CID 86716613.
- ^ Fisher 1930
- ^ a b c Yampolsky, Lev Y.; Stoltzfus, Arlin (20 December 2001). "Bias in the introduction of variation as an orienting factor in evolution". Evolution & Development. 3 (2): 73–83. doi:10.1046/j.1525-142x.2001.003002073.x. PMID 11341676. S2CID 26956345.
- ^ Sueoka, Noboru (1 April 1962). "On the Genetic Basis of Variation and Heterogeneity of DNA Base Composition". PNAS. 48 (4): 582–592. Bibcode:1962PNAS...48..582S. doi:10.1073/pnas.48.4.582. PMC 220819. PMID 13918161.
- ^ Freese, Ernst (July 1962). "On the Evolution of the Base Composition of DNA". Journal of Theoretical Biology. 3 (1): 82–101. Bibcode:1962JThBi...3...82F. doi:10.1016/S0022-5193(62)80005-8.
- ^ Cox, Edward C.; Yanofsky, Charles (1 November 1967). "Altered base ratios in the DNA of an Escherichia coli mutator strain". Proc. Natl. Acad. Sci. USA. 58 (5): 1895–1902. Bibcode:1967PNAS...58.1895C. doi:10.1073/pnas.58.5.1895. PMC 223881. PMID 4866980.
- ^ Shah, Premal; Gilchrist, Michael A. (21 June 2011). "Explaining complex codon usage patterns with selection for translational efficiency, mutation bias, and genetic drift". PNAS. 108 (25): 10231–10236. Bibcode:2011PNAS..10810231S. doi:10.1073/pnas.1016719108. PMC 3121864. PMID 21646514.
- ^ Bulmer, Michael G. (November 1991). "The selection-mutation-drift theory of synonymous codon usage". Genetics. 129 (3): 897–907. doi:10.1093/genetics/129.3.897. PMC 1204756. PMID 1752426.
- ^ Fryxell, Karl J.; Zuckerkandl, Emile (September 2000). "Cytosine Deamination Plays a Primary Role in the Evolution of Mammalian Isochores". Molecular Biology and Evolution. 17 (9): 1371–1383. doi:10.1093/oxfordjournals.molbev.a026420. PMID 10958853.
- ^ Petrov, Dmitri A.; Sangster, Todd A.; Johnston, J. Spencer; et al. (11 February 2000). "Evidence for DNA Loss as a Determinant of Genome Size". Science. 287 (5455): 1060–1062. Bibcode:2000Sci...287.1060P. doi:10.1126/science.287.5455.1060. ISSN 0036-8075. PMID 10669421. S2CID 12021662.
- ^ Petrov, Dmitri A. (May 2002). "DNA loss and evolution of genome size in Drosophila". Genetica. 115 (1): 81–91. doi:10.1023/A:1016076215168. ISSN 0016-6707. PMID 12188050. S2CID 5314242.
- ^ Duret, Laurent; Galtier, Nicolas (September 2009). "Biased Gene Conversion and the Evolution of Mammalian Genomic Landscapes". Annual Review of Genomics and Human Genetics. 10. Annual Reviews: 285–311. doi:10.1146/annurev-genom-082908-150001. PMID 19630562. S2CID 9126286.
- ^ Hershberg, Ruth; Petrov, Dmitri A. (9 September 2010). "Evidence That Mutation Is Universally Biased towards AT in Bacteria". PLOS Genetics. 6 (9): e1001115. doi:10.1371/journal.pgen.1001115. PMC 2936535. PMID 20838599.
- ^ A. Stoltzfus (2019). "Understanding bias in the introduction of variation as an evolutionary cause". In Uller, T.; Laland, K.N. (eds.). Evolutionary Causation: Biological and Philosophical Reflections. Cambridge, MA: MIT Press.
- ^ Stoltzfus, Arlin; McCandlish, David M. (September 2017). "Mutational Biases Influence Parallel Adaptation". Molecular Biology and Evolution. 34 (9): 2163–2172. doi:10.1093/molbev/msx180. PMC 5850294. PMID 28645195.
- ^ Payne, Joshua L.; Menardo, Fabrizio; Trauner, Andrej; et al. (13 May 2019). "Transition bias influences the evolution of antibiotic resistance in Mycobacterium tuberculosis". PLOS Biology. 17 (5): e3000265. doi:10.1371/journal.pbio.3000265. PMC 6532934. PMID 31083647.
- ^ Storz, Jay F.; Natarajan, Chandrasekhar; Signore, Anthony V.; et al. (22 July 2019). "The role of mutation bias in adaptive molecular evolution: insights from convergent changes in protein function". Philosophical Transactions of the Royal Society B. 374 (1777): 20180238. doi:10.1098/rstb.2018.0238. PMC 6560279. PMID 31154983.
- ^ Svensson, Erik I.; Berger, David (1 May 2019). "The Role of Mutation Bias in Adaptive Evolution". Trends in Ecology & Evolution. 34 (5): 422–434. Bibcode:2019TEcoE..34..422S. doi:10.1016/j.tree.2019.01.015. PMID 31003616. S2CID 125066709.
- ^ Lien, Sigbjørn; Szyda, Joanna; Schechinger, Birgit; et al. (February 2000). "Evidence for Heterogeneity in Recombination in the Human Pseudoautosomal Region: High Resolution Analysis by Sperm Typing and Radiation-Hybrid Mapping". American Journal of Human Genetics. 66 (2): 557–566. doi:10.1086/302754. ISSN 0002-9297. PMC 1288109. PMID 10677316.
- ^ Barton, Nicholas H. (29 November 2000). "Genetic hitchhiking". Philosophical Transactions of the Royal Society B. 355 (1403): 1553–1562. doi:10.1098/rstb.2000.0716. ISSN 0962-8436. PMC 1692896. PMID 11127900.
- ^ Ries, C; Spaethe, J; Sztatecsny, M; Strondl, C; Hödl, W (20 October 2008). "Turning blue and ultraviolet: sex-specific colour change during the mating season in the Balkan moor frog". Journal of Zoology. 276 (3): 229–236. doi:10.1111/j.1469-7998.2008.00456.x – via Google Scholar.
- ^ Andersson, Malte; Simmons, Leigh W. (June 2006). "Sexual selection and mate choice" (PDF). Trends in Ecology & Evolution. 21 (6): 296–302. CiteSeerX 10.1.1.595.4050. doi:10.1016/j.tree.2006.03.015. ISSN 0169-5347. PMID 16769428. Archived (PDF) from the original on 9 March 2013.
- ^ Kokko, Hanna; Brooks, Robert; McNamara, John M.; Houston, Alasdair I. (7 July 2002). "The sexual selection continuum". Proceedings of the Royal Society B. 269 (1498): 1331–1340. doi:10.1098/rspb.2002.2020. ISSN 0962-8452. PMC 1691039. PMID 12079655.
- ^ Quinn, Thomas P.; Hendry, Andrew P.; Buck, Gregory B. (2001). "Balancing natural and sexual selection in sockeye salmon: interactions between body size, reproductive opportunity and vulnerability to predation by bears" (PDF). Evolutionary Ecology Research. 3: 917–937. ISSN 1522-0613. Archived (PDF) from the original on 5 March 2016. Retrieved 15 December 2014.
- ^ Hunt, John; Brooks, Robert; Jennions, Michael D.; et al. (23 December 2004). "High-quality male field crickets invest heavily in sexual display but die young". Nature. 432 (7020): 1024–1027. Bibcode:2004Natur.432.1024H. doi:10.1038/nature03084. ISSN 0028-0836. PMID 15616562. S2CID 4417867.
- ^ Baym, Michael; Lieberman, Tami D.; Kelsic, Eric D.; et al. (9 September 2016). "Spatiotemporal microbial evolution on antibiotic landscapes". Science. 353 (6304): 1147–1151. Bibcode:2016Sci...353.1147B. doi:10.1126/science.aag0822. ISSN 0036-8075. PMC 5534434. PMID 27609891.
- ^ a b c Scott, Eugenie C.; Matzke, Nicholas J. (15 May 2007). "Biological design in science classrooms". PNAS. 104 (Suppl. 1): 8669–8676. Bibcode:2007PNAS..104.8669S. doi:10.1073/pnas.0701505104. PMC 1876445. PMID 17494747.
- ^ Hendry, Andrew Paul; Kinnison, Michael T. (November 2001). "An introduction to microevolution: rate, pattern, process". Genetica. 112–113 (1): 1–8. doi:10.1023/A:1013368628607. ISSN 0016-6707. PMID 11838760. S2CID 24485535.
- ^ Leroi, Armand M. (March–April 2000). "The scale independence of evolution". Evolution & Development. 2 (2): 67–77. CiteSeerX 10.1.1.120.1020. doi:10.1046/j.1525-142x.2000.00044.x. ISSN 1520-541X. PMID 11258392. S2CID 17289010.
- ^ Gould 2002, pp. 657–658.
- ^ a b Gould, Stephen Jay (19 July 1994). "Tempo and mode in the macroevolutionary reconstruction of Darwinism". PNAS. 91 (15): 6764–6771. Bibcode:1994PNAS...91.6764G. doi:10.1073/pnas.91.15.6764. PMC 44281. PMID 8041695.
- ^ Jablonski, David (2000). "Micro- and macroevolution: scale and hierarchy in evolutionary biology and paleobiology". Paleobiology. 26 (sp4): 15–52. doi:10.1666/0094-8373(2000)26[15:MAMSAH]2.0.CO;2. S2CID 53451360.
- ^ Dougherty, Michael J. (20 July 1998). "Is the human race evolving or devolving?". Scientific American. ISSN 0036-8733. Archived from the original on 6 May 2014. Retrieved 11 September 2015.
- ^ Isaak, Mark, ed. (22 July 2003). "Claim CB932: Evolution of degenerate forms". TalkOrigins Archive. Houston, Texas: The TalkOrigins Foundation. Archived from the original on 23 August 2014. Retrieved 19 December 2014.
- ^ Lane 1996, p. 61
- ^ Carroll, Sean B. (22 February 2001). "Chance and necessity: the evolution of morphological complexity and diversity". Nature. 409 (6823): 1102–1109. Bibcode:2001Natur.409.1102C. doi:10.1038/35059227. PMID 11234024. S2CID 4319886.
- ^ Whitman, William B.; Coleman, David C.; Wiebe, William J. (9 June 1998). "Prokaryotes: The unseen majority". PNAS. 95 (12): 6578–6583. Bibcode:1998PNAS...95.6578W. doi:10.1073/pnas.95.12.6578. ISSN 0027-8424. PMC 33863. PMID 9618454.
- ^ a b Schloss, Patrick D.; Handelsman, Jo (December 2004). "Status of the Microbial Census". Microbiology and Molecular Biology Reviews. 68 (4): 686–691. doi:10.1128/MMBR.68.4.686-691.2004. PMC 539005. PMID 15590780.
- ^ Nealson, Kenneth H. (January 1999). "Post-Viking microbiology: new approaches, new data, new insights". Origins of Life and Evolution of Biospheres. 29 (1): 73–93. Bibcode:1999OLEB...29...73N. doi:10.1023/A:1006515817767. ISSN 0169-6149. PMID 11536899. S2CID 12289639.
- ^ Buckling, Angus; MacLean, R. Craig; Brockhurst, Michael A.; Colegrave, Nick (12 February 2009). "The Beagle in a bottle". Nature. 457 (7231): 824–829. Bibcode:2009Natur.457..824B. doi:10.1038/nature07892. ISSN 0028-0836. PMID 19212400. S2CID 205216404.
- ^ Elena, Santiago F.; Lenski, Richard E. (June 2003). "Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation". Nature Reviews Genetics. 4 (6): 457–469. doi:10.1038/nrg1088. ISSN 1471-0056. PMID 12776215. S2CID 209727.
- ^ Mayr 1982, p. 483: "Adaptation... could no longer be considered a static condition, a product of a creative past and became instead a continuing dynamic process."
- ^ The sixth edition of the Oxford Dictionary of Science (2010) defines adaptation as "Any change in the structure or functioning of successive generations of a population that makes it better suited to its environment."
- ^ Orr, H. Allen (February 2005). "The genetic theory of adaptation: a brief history". Nature Reviews Genetics. 6 (2): 119–127. doi:10.1038/nrg1523. ISSN 1471-0056. PMID 15716908. S2CID 17772950.
- ^ Dobzhansky 1968, pp. 1–34
- ^ Dobzhansky 1970, pp. 4–6, 79–82, 84–87
- ^ Dobzhansky, Theodosius (March 1956). "Genetics of Natural Populations. XXV. Genetic Changes in Populations of Drosophila pseudoobscura and Drosophila persimilis in Some Localities in California". Evolution. 10 (1): 82–92. doi:10.2307/2406099. ISSN 0014-3820. JSTOR 2406099.
- ^ Nakajima, Akira; Sugimoto, Yohko; Yoneyama, Hiroshi; et al. (June 2002). "High-Level Fluoroquinolone Resistance in Pseudomonas aeruginosa Due to Interplay of the MexAB-OprM Efflux Pump and the DNA Gyrase Mutation". Microbiology and Immunology. 46 (6): 391–395. doi:10.1111/j.1348-0421.2002.tb02711.x. ISSN 1348-0421. PMID 12153116. S2CID 22593331.
- ^ Blount, Zachary D.; Borland, Christina Z.; Lenski, Richard E. (10 June 2008). "Inaugural Article: Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli". PNAS. 105 (23): 7899–7906. Bibcode:2008PNAS..105.7899B. doi:10.1073/pnas.0803151105. ISSN 0027-8424. PMC 2430337. PMID 18524956.
- ^ Okada, Hirosuke; Negoro, Seiji; Kimura, Hiroyuki; et al. (10 November 1983). "Evolutionary adaptation of plasmid-encoded enzymes for degrading nylon oligomers". Nature. 306 (5939): 203–206. Bibcode:1983Natur.306..203O. doi:10.1038/306203a0. ISSN 0028-0836. PMID 6646204. S2CID 4364682.
- ^ Ohno, Susumu (April 1984). "Birth of a unique enzyme from an alternative reading frame of the preexisted, internally repetitious coding sequence". PNAS. 81 (8): 2421–2425. Bibcode:1984PNAS...81.2421O. doi:10.1073/pnas.81.8.2421. ISSN 0027-8424. PMC 345072. PMID 6585807.
- ^ Copley, Shelley D. (June 2000). "Evolution of a metabolic pathway for degradation of a toxic xenobiotic: the patchwork approach". Trends in Biochemical Sciences. 25 (6): 261–265. doi:10.1016/S0968-0004(00)01562-0. ISSN 0968-0004. PMID 10838562.
- ^ Crawford, Ronald L.; Jung, Carina M.; Strap, Janice L. (October 2007). "The recent evolution of pentachlorophenol (PCP)-4-monooxygenase (PcpB) and associated pathways for bacterial degradation of PCP". Biodegradation. 18 (5): 525–539. doi:10.1007/s10532-006-9090-6. ISSN 0923-9820. PMID 17123025. S2CID 8174462.
- ^ Altenberg 1995, pp. 205–259
- ^ Masel, Joanna; Bergman, Aviv (July 2003). "The evolution of the evolvability properties of the yeast prion [PSI+]". Evolution. 57 (7): 1498–1512. doi:10.1111/j.0014-3820.2003.tb00358.x. PMID 12940355. S2CID 30954684.
- ^ Lancaster, Alex K.; Bardill, J. Patrick; True, Heather L.; Masel, Joanna (February 2010). "The Spontaneous Appearance Rate of the Yeast Prion [PSI+] and Its Implications for the Evolution of the Evolvability Properties of the [PSI+] System". Genetics. 184 (2): 393–400. doi:10.1534/genetics.109.110213. ISSN 0016-6731. PMC 2828720. PMID 19917766.
- ^ Draghi, Jeremy; Wagner, Günter P. (February 2008). "Evolution of evolvability in a developmental model". Evolution. 62 (2): 301–315. doi:10.1111/j.1558-5646.2007.00303.x. PMID 18031304. S2CID 11560256.
- ^ a b Bejder, Lars; Hall, Brian K. (November 2002). "Limbs in whales and limblessness in other vertebrates: mechanisms of evolutionary and developmental transformation and loss". Evolution & Development. 4 (6): 445–458. doi:10.1046/j.1525-142X.2002.02033.x. PMID 12492145. S2CID 8448387.
- ^ Young, Nathan M.; HallgrÍmsson, Benedikt (December 2005). "Serial homology and the evolution of mammalian limb covariation structure". Evolution. 59 (12): 2691–2704. doi:10.1554/05-233.1. ISSN 0014-3820. PMID 16526515. S2CID 198156135.
- ^ a b Penny, David; Poole, Anthony (December 1999). "The nature of the last universal common ancestor". Current Opinion in Genetics & Development. 9 (6): 672–677. doi:10.1016/S0959-437X(99)00020-9. PMID 10607605.
- ^ Hall, Brian K. (August 2003). "Descent with modification: the unity underlying homology and homoplasy as seen through an analysis of development and evolution". Biological Reviews. 78 (3): 409–433. doi:10.1017/S1464793102006097. ISSN 1464-7931. PMID 14558591. S2CID 22142786.
- ^ Shubin, Neil; Tabin, Clifford J.; Carroll, Sean B. (12 February 2009). "Deep homology and the origins of evolutionary novelty". Nature. 457 (7231): 818–823. Bibcode:2009Natur.457..818S. doi:10.1038/nature07891. PMID 19212399. S2CID 205216390.
- ^ a b c Fong, Daniel F.; Kane, Thomas C.; Culver, David C. (November 1995). "Vestigialization and Loss of Nonfunctional Characters". Annual Review of Ecology and Systematics. 26 (1): 249–268. Bibcode:1995AnRES..26..249F. doi:10.1146/annurev.es.26.110195.001341.
- ^ ZhaoLei Zhang; Gerstein, Mark (August 2004). "Large-scale analysis of pseudogenes in the human genome". Current Opinion in Genetics & Development. 14 (4): 328–335. doi:10.1016/j.gde.2004.06.003. ISSN 0959-437X. PMID 15261647.
- ^ Jeffery, William R. (May–June 2005). "Adaptive Evolution of Eye Degeneration in the Mexican Blind Cavefish". Journal of Heredity. 96 (3): 185–196. CiteSeerX 10.1.1.572.6605. doi:10.1093/jhered/esi028. PMID 15653557.
- ^ Maxwell, Erin E.; Larsson, Hans C.E. (May 2007). "Osteology and myology of the wing of the Emu (Dromaius novaehollandiae) and its bearing on the evolution of vestigial structures". Journal of Morphology. 268 (5): 423–441. doi:10.1002/jmor.10527. ISSN 0362-2525. PMID 17390336. S2CID 12494187.
- ^ van der Kooi, Casper J.; Schwander, Tanja (November 2014). "On the fate of sexual traits under asexuality" (PDF). Biological Reviews. 89 (4): 805–819. doi:10.1111/brv.12078. ISSN 1464-7931. PMID 24443922. S2CID 33644494. Archived (PDF) from the original on 23 July 2015. Retrieved 5 August 2015.
- ^ Silvestri, Anthony R. Jr.; Singh, Iqbal (April 2003). "The unresolved problem of the third molar: Would people be better off without it?". Journal of the American Dental Association. 134 (4): 450–455. doi:10.14219/jada.archive.2003.0194. PMID 12733778. Archived from the original on 23 August 2014.
- ^ Coyne 2009, p. 62
- ^ Darwin 1872, pp. 101, 103
- ^ Gray 2007, p. 66
- ^ Coyne 2009, pp. 85–86
- ^ Stevens 1982, p. 87
- ^ a b Gould 2002, pp. 1235–1236.
- ^ Pallen, Mark J.; Matzke, Nicholas J. (October 2006). "From The Origin of Species to the origin of bacterial flagella" (PDF). Nature Reviews Microbiology (PDF). 4 (10): 784–790. doi:10.1038/nrmicro1493. ISSN 1740-1526. PMID 16953248. S2CID 24057949. Archived from the original (PDF) on 26 December 2014. Retrieved 25 December 2014.
- ^ Clements, Abigail; Bursac, Dejan; Gatsos, Xenia; et al. (15 September 2009). "The reducible complexity of a mitochondrial molecular machine". PNAS. 106 (37): 15791–15795. Bibcode:2009PNAS..10615791C. doi:10.1073/pnas.0908264106. PMC 2747197. PMID 19717453.
- ^ Piatigorsky et al. 1994, pp. 241–250
- ^ Wistow, Graeme (August 1993). "Lens crystallins: gene recruitment and evolutionary dynamism". Trends in Biochemical Sciences. 18 (8): 301–306. doi:10.1016/0968-0004(93)90041-K. ISSN 0968-0004. PMID 8236445.
- ^ Johnson, Norman A.; Porter, Adam H. (November 2001). "Toward a new synthesis: population genetics and evolutionary developmental biology". Genetica. 112–113 (1): 45–58. doi:10.1023/A:1013371201773. ISSN 0016-6707. PMID 11838782. S2CID 1651351.
- ^ Baguñà, Jaume; Garcia-Fernàndez, Jordi (2003). "Evo-Devo: the long and winding road". The International Journal of Developmental Biology. 47 (7–8): 705–713. ISSN 0214-6282. PMID 14756346. Archived from the original on 28 November 2014.
- Love, Alan C. (March 2003). "Evolutionary Morphology, Innovation and the Synthesis of Evolutionary and Developmental Biology". Biology and Philosophy. 18 (2): 309–345. doi:10.1023/A:1023940220348. S2CID 82307503.
- ^ Allin, Edgar F. (December 1975). "Evolution of the mammalian middle ear". Journal of Morphology. 147 (4): 403–437. doi:10.1002/jmor.1051470404. ISSN 0362-2525. PMID 1202224. S2CID 25886311.
- ^ Harris, Matthew P.; Hasso, Sean M.; Ferguson, Mark W.J.; Fallon, John F. (21 February 2006). "The Development of Archosaurian First-Generation Teeth in a Chicken Mutant". Current Biology. 16 (4): 371–377. Bibcode:2006CBio...16..371H. doi:10.1016/j.cub.2005.12.047. PMID 16488870. S2CID 15733491.
- ^ Carroll, Sean B. (11 July 2008). "Evo-Devo and an Expanding Evolutionary Synthesis: A Genetic Theory of Morphological Evolution". Cell. 134 (1): 25–36. doi:10.1016/j.cell.2008.06.030. PMID 18614008. S2CID 2513041.
- ^ Wade, Michael J. (March 2007). "The co-evolutionary genetics of ecological communities". Nature Reviews Genetics. 8 (3): 185–195. doi:10.1038/nrg2031. PMID 17279094. S2CID 36705246.
- ^ Geffeney, Shana; Brodie, Edmund D. Jr.; Ruben, Peter C.; Brodie, Edmund D. III (23 August 2002). "Mechanisms of Adaptation in a Predator-Prey Arms Race: TTX-Resistant Sodium Channels". Science. 297 (5585): 1336–1339. Bibcode:2002Sci...297.1336G. doi:10.1126/science.1074310. PMID 12193784. S2CID 8816337.
- Brodie, Edmund D. Jr.; Ridenhour, Benjamin J.; Brodie, Edmund D. III (October 2002). "The evolutionary response of predators to dangerous prey: hotspots and coldspots in the geographic mosaic of coevolution between garter snakes and newts". Evolution. 56 (10): 2067–2082. doi:10.1554/0014-3820(2002)056[2067:teropt]2.0.co;2. PMID 12449493. S2CID 8251443.
- Carroll, Sean B. (21 December 2009). "Whatever Doesn't Kill Some Animals Can Make Them Deadly". The New York Times. New York. Archived from the original on 23 April 2015. Retrieved 26 December 2014.
- ^ Sachs, Joel L. (September 2006). "Cooperation within and among species". Journal of Evolutionary Biology. 19 (5): 1415–1418, discussion 1426–1436. doi:10.1111/j.1420-9101.2006.01152.x. PMID 16910971. S2CID 4828678.
- Nowak, Martin A. (8 December 2006). "Five Rules for the Evolution of Cooperation". Science. 314 (5805): 1560–1563. Bibcode:2006Sci...314.1560N. doi:10.1126/science.1133755. PMC 3279745. PMID 17158317.
- ^ Paszkowski, Uta (August 2006). "Mutualism and parasitism: the yin and yang of plant symbioses". Current Opinion in Plant Biology. 9 (4): 364–370. Bibcode:2006COPB....9..364P. doi:10.1016/j.pbi.2006.05.008. ISSN 1369-5266. PMID 16713732.
- ^ Hause, Bettina; Fester, Thomas (May 2005). "Molecular and cell biology of arbuscular mycorrhizal symbiosis". Planta. 221 (2): 184–196. Bibcode:2005Plant.221..184H. doi:10.1007/s00425-004-1436-x. PMID 15871030. S2CID 20082902.
- ^ Bertram, John S. (December 2000). "The molecular biology of cancer". Molecular Aspects of Medicine. 21 (6): 167–223. doi:10.1016/S0098-2997(00)00007-8. PMID 11173079. S2CID 24155688.
- ^ Reeve, H. Kern; Hölldobler, Bert (5 June 2007). "The emergence of a superorganism through intergroup competition". PNAS. 104 (23): 9736–9740. Bibcode:2007PNAS..104.9736R. doi:10.1073/pnas.0703466104. ISSN 0027-8424. PMC 1887545. PMID 17517608.
- ^ Axelrod, Robert; Hamilton, W. D. (27 March 1981). "The evolution of cooperation". Science. 211 (4489): 1390–1396. Bibcode:1981Sci...211.1390A. doi:10.1126/science.7466396. PMID 7466396.
- ^ Wilson, Edward O.; Hölldobler, Bert (20 September 2005). "Eusociality: Origin and consequences". PNAS. 102 (38): 13367–1371. Bibcode:2005PNAS..10213367W. doi:10.1073/pnas.0505858102. PMC 1224642. PMID 16157878.
- ^ a b Gavrilets, Sergey (October 2003). "Perspective: models of speciation: what have we learned in 40 years?". Evolution. 57 (10): 2197–2215. doi:10.1554/02-727. PMID 14628909. S2CID 198158082.
- ^ a b c de Queiroz, Kevin (3 May 2005). "Ernst Mayr and the modern concept of species". PNAS. 102 (Suppl. 1): 6600–6607. Bibcode:2005PNAS..102.6600D. doi:10.1073/pnas.0502030102. PMC 1131873. PMID 15851674.
- ^ a b Ereshefsky, Marc (December 1992). "Eliminative pluralism". Philosophy of Science. 59 (4): 671–690. doi:10.1086/289701. JSTOR 188136. S2CID 224829314.
- ^ Mayr 1942, p. 120
- ^ Fraser, Christophe; Alm, Eric J.; Polz, Martin F.; et al. (6 February 2009). "The Bacterial Species Challenge: Making Sense of Genetic and Ecological Diversity". Science. 323 (5915): 741–746. Bibcode:2009Sci...323..741F. doi:10.1126/science.1159388. PMID 19197054. S2CID 15763831.
- ^ Short, Roger Valentine (October 1975). "The contribution of the mule to scientific thought". Journal of Reproduction and Fertility. Supplement (23): 359–364. OCLC 1639439. PMID 1107543.
- ^ Gross, Briana L.; Rieseberg, Loren H. (May–June 2005). "The Ecological Genetics of Homoploid Hybrid Speciation". Journal of Heredity. 96 (3): 241–252. doi:10.1093/jhered/esi026. ISSN 0022-1503. PMC 2517139. PMID 15618301.
- ^ Burke, John M.; Arnold, Michael L. (December 2001). "Genetics and the fitness of hybrids". Annual Review of Genetics. 35: 31–52. doi:10.1146/annurev.genet.35.102401.085719. ISSN 0066-4197. PMID 11700276. S2CID 26683922.
- ^ Vrijenhoek, Robert C. (4 April 2006). "Polyploid Hybrids: Multiple Origins of a Treefrog Species". Current Biology. 16 (7): R245–R247. Bibcode:2006CBio...16.R245V. doi:10.1016/j.cub.2006.03.005. ISSN 0960-9822. PMID 16581499. S2CID 11657663.
- ^ Rice, William R.; Hostert, Ellen E. (December 1993). "Laboratory Experiments on Speciation: What Have We Learned in 40 Years?". Evolution. 47 (6): 1637–1653. doi:10.1111/j.1558-5646.1993.tb01257.x. ISSN 0014-3820. JSTOR 2410209. PMID 28568007. S2CID 42100751.
- Jiggins, Chris D.; Bridle, Jon R. (March 2004). "Speciation in the apple maggot fly: a blend of vintages?". Trends in Ecology & Evolution. 19 (3): 111–114. doi:10.1016/j.tree.2003.12.008. ISSN 0169-5347. PMID 16701238.
- Boxhorn, Joseph (1 September 1995). "Observed Instances of Speciation". TalkOrigins Archive. Houston, Texas: The TalkOrigins Foundation, Inc. Archived from the original on 22 January 2009. Retrieved 26 December 2008.
- Weinberg, James R.; Starczak, Victoria R.; Jörg, Daniele (August 1992). "Evidence for Rapid Speciation Following a Founder Event in the Laboratory". Evolution. 46 (4): 1214–1220. doi:10.2307/2409766. ISSN 0014-3820. JSTOR 2409766. PMID 28564398.
- ^ Herrel, Anthony; Huyghe, Katleen; Vanhooydonck, Bieke; et al. (25 March 2008). "Rapid large-scale evolutionary divergence in morphology and performance associated with exploitation of a different dietary resource". PNAS. 105 (12): 4792–4795. Bibcode:2008PNAS..105.4792H. doi:10.1073/pnas.0711998105. ISSN 0027-8424. PMC 2290806. PMID 18344323.
- ^ Losos, Jonathan B.; Warhelt, Kenneth I.; Schoener, Thomas W. (1 May 1997). "Adaptive differentiation following experimental island colonization in Anolis lizards". Nature. 387 (6628): 70–73. Bibcode:1997Natur.387...70L. doi:10.1038/387070a0. ISSN 0028-0836. S2CID 4242248.
- ^ Hoskin, Conrad J.; Higgle, Megan; McDonald, Keith R.; Moritz, Craig (27 October 2005). "Reinforcement drives rapid allopatric speciation". Nature. 437 (7063): 1353–1356. Bibcode:2005Natur.437.1353H. doi:10.1038/nature04004. PMID 16251964. S2CID 4417281.
- ^ Templeton, Alan R. (April 1980). "The Theory of Speciation VIA the Founder Principle". Genetics. 94 (4): 1011–1038. doi:10.1093/genetics/94.4.1011. PMC 1214177. PMID 6777243. Archived from the original on 23 August 2014. Retrieved 29 December 2014.
- ^ Antonovics, Janis (July 2006). "Evolution in closely adjacent plant populations X: long-term persistence of prereproductive isolation at a mine boundary". Heredity. 97 (1): 33–37. doi:10.1038/sj.hdy.6800835. ISSN 0018-067X. PMID 16639420. S2CID 12291411.
- ^ Nosil, Patrik; Crespi, Bernard J.; Gries, Regine; Gries, Gerhard (March 2007). "Natural selection and divergence in mate preference during speciation". Genetica. 129 (3): 309–327. doi:10.1007/s10709-006-0013-6. ISSN 0016-6707. PMID 16900317. S2CID 10808041.
- ^ Savolainen, Vincent; Anstett, Marie-Charlotte; Lexer, Christian; et al. (11 May 2006). "Sympatric speciation in palms on an oceanic island". Nature. 441 (7090): 210–213. Bibcode:2006Natur.441..210S. doi:10.1038/nature04566. ISSN 0028-0836. PMID 16467788. S2CID 867216.
- Barluenga, Marta; Stölting, Kai N.; Salzburger, Walter; et al. (9 February 2006). "Sympatric speciation in Nicaraguan crater lake cichlid fish". Nature. 439 (7077): 719–23. Bibcode:2006Natur.439..719B. doi:10.1038/nature04325. ISSN 0028-0836. PMID 16467837. S2CID 3165729. Archived from the original on 30 July 2022. Retrieved 30 July 2022.
- ^ Gavrilets, Sergey (21 March 2006). "The Maynard Smith model of sympatric speciation". Journal of Theoretical Biology. 239 (2): 172–182. Bibcode:2006JThBi.239..172G. doi:10.1016/j.jtbi.2005.08.041. ISSN 0022-5193. PMID 16242727.
- ^ Wood, Troy E.; Takebayashi, Naoki; Barker, Michael S.; et al. (18 August 2009). "The frequency of polyploid speciation in vascular plants". PNAS. 106 (33): 13875–13879. Bibcode:2009PNAS..10613875W. doi:10.1073/pnas.0811575106. ISSN 0027-8424. PMC 2728988. PMID 19667210.
- ^ Hegarty, Matthew J.; Hiscock, Simon J. (20 May 2008). "Genomic Clues to the Evolutionary Success of Polyploid Plants". Current Biology. 18 (10): R435–R444. Bibcode:2008CBio...18.R435H. doi:10.1016/j.cub.2008.03.043. ISSN 0960-9822. PMID 18492478. S2CID 1584282.
- ^ Jakobsson, Mattias; Hagenblad, Jenny; Tavaré, Simon; et al. (June 2006). "A Unique Recent Origin of the Allotetraploid Species Arabidopsis suecica: Evidence from Nuclear DNA Markers". Molecular Biology and Evolution. 23 (6): 1217–1231. doi:10.1093/molbev/msk006. PMID 16549398. Archived from the original on 15 February 2022. Retrieved 30 July 2022.
- ^ Säll, Torbjörn; Jakobsson, Mattias; Lind-Halldén, Christina; Halldén, Christer (September 2003). "Chloroplast DNA indicates a single origin of the allotetraploid Arabidopsis suecica". Journal of Evolutionary Biology. 16 (5): 1019–1029. doi:10.1046/j.1420-9101.2003.00554.x. PMID 14635917. S2CID 29281998.
- ^ Bomblies, Kirsten; Weigel, Detlef (December 2007). "Arabidopsis—a model genus for speciation". Current Opinion in Genetics & Development. 17 (6): 500–504. doi:10.1016/j.gde.2007.09.006. PMID 18006296.
- ^ Sémon, Marie; Wolfe, Kenneth H. (December 2007). "Consequences of genome duplication". Current Opinion in Genetics & Development. 17 (6): 505–512. doi:10.1016/j.gde.2007.09.007. PMID 18006297.
- ^ Eldredge & Gould 1972, pp. 82–115
- ^ Benton, Michael J. (7 April 1995). "Diversification and extinction in the history of life". Science. 268 (5207): 52–58. Bibcode:1995Sci...268...52B. doi:10.1126/science.7701342. ISSN 0036-8075. PMID 7701342.
- ^ Raup, David M. (28 March 1986). "Biological extinction in Earth history". Science. 231 (4745): 1528–1533. Bibcode:1986Sci...231.1528R. doi:10.1126/science.11542058. PMID 11542058. S2CID 23012011.
- ^ Avise, John C.; Hubbell, Stephen P.; Ayala, Francisco J. (12 August 2008). "In the light of evolution II: Biodiversity and extinction". PNAS. 105 (Suppl. 1): 11453–11457. Bibcode:2008PNAS..10511453A. doi:10.1073/pnas.0802504105. PMC 2556414. PMID 18695213.
- ^ a b c Raup, David M. (19 July 1994). "The role of extinction in evolution". PNAS. 91 (15): 6758–6763. Bibcode:1994PNAS...91.6758R. doi:10.1073/pnas.91.15.6758. PMC 44280. PMID 8041694.
- ^ Novacek, Michael J.; Cleland, Elsa E. (8 May 2001). "The current biodiversity extinction event: scenarios for mitigation and recovery". PNAS. 98 (10): 5466–5470. Bibcode:2001PNAS...98.5466N. doi:10.1073/pnas.091093698. ISSN 0027-8424. PMC 33235. PMID 11344295.
- ^ Pimm, Stuart; Raven, Peter; Peterson, Alan; et al. (18 July 2006). "Human impacts on the rates of recent, present and future bird extinctions". PNAS. 103 (29): 10941–10946. Bibcode:2006PNAS..10310941P. doi:10.1073/pnas.0604181103. ISSN 0027-8424. PMC 1544153. PMID 16829570.
- ^ Barnosky, Anthony D.; Koch, Paul L.; Feranec, Robert S.; et al. (1 October 2004). "Assessing the Causes of Late Pleistocene Extinctions on the Continents". Science. 306 (5693): 70–75. Bibcode:2004Sci...306...70B. CiteSeerX 10.1.1.574.332. doi:10.1126/science.1101476. ISSN 0036-8075. PMID 15459379. S2CID 36156087.
- ^ Lewis, Owen T. (29 January 2006). "Climate change, species–area curves and the extinction crisis". Philosophical Transactions of the Royal Society B. 361 (1465): 163–171. doi:10.1098/rstb.2005.1712. ISSN 0962-8436. PMC 1831839. PMID 16553315.
- ^ a b Stearns & Stearns 1999, p. X
- ^ a b Novacek, Michael J. (8 November 2014). "Prehistory's Brilliant Future". The New York Times. New York. ISSN 0362-4331. Archived from the original on 29 December 2014. Retrieved 25 December 2014.
- ^ "Researchers find that Earth may be home to 1 trillion species". National Science Foundation. Arlington County, Virginia. 2 May 2016. Archived from the original on 4 May 2016. Retrieved 6 May 2016.
- ^ a b c Kutschera, Ulrich; Niklas, Karl J. (June 2004). "The modern theory of biological evolution: an expanded synthesis". Naturwissenschaften. 91 (6): 255–276. Bibcode:2004NW.....91..255K. doi:10.1007/s00114-004-0515-y. ISSN 1432-1904. PMID 15241603. S2CID 10731711.
- ^ Jablonski, David (8 May 2001). "Lessons from the past: Evolutionary impacts of mass extinctions". PNAS. 98 (10): 5393–5398. Bibcode:2001PNAS...98.5393J. doi:10.1073/pnas.101092598. PMC 33224. PMID 11344284.
- ^ Bull, James J.; Wichman, Holly A. (November 2001). "Applied evolution". Annual Review of Ecology and Systematics. 32 (1): 183–217. Bibcode:2001AnRES..32..183B. doi:10.1146/annurev.ecolsys.32.081501.114020. ISSN 1545-2069.
- ^ Doebley, John F.; Gaut, Brandon S.; Smith, Bruce D. (29 December 2006). "The Molecular Genetics of Crop Domestication". Cell. 127 (7): 1309–1321. doi:10.1016/j.cell.2006.12.006. ISSN 0092-8674. PMID 17190597. S2CID 278993.
- ^ Jäckel, Christian; Kast, Peter; Hilvert, Donald (June 2008). "Protein Design by Directed Evolution". Annual Review of Biophysics. 37: 153–173. doi:10.1146/annurev.biophys.37.032807.125832. ISSN 1936-122X. PMID 18573077.
- ^ Maher, Brendan (8 April 2009). "Evolution: Biology's next top model?". Nature. 458 (7239): 695–698. doi:10.1038/458695a. ISSN 0028-0836. PMID 19360058. S2CID 41648315.
- ^ Borowsky, Richard (8 January 2008). "Restoring sight in blind cavefish". Current Biology. 18 (1): R23–R24. Bibcode:2008CBio...18..R23B. doi:10.1016/j.cub.2007.11.023. ISSN 0960-9822. PMID 18177707. S2CID 16967690.
- ^ Gross, Joshua B.; Borowsky, Richard; Tabin, Clifford J. (2 January 2009). Barsh, Gregory S. (ed.). "A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus". PLOS Genetics. 5 (1): e1000326. doi:10.1371/journal.pgen.1000326. ISSN 1553-7390. PMC 2603666. PMID 19119422.
- ^ Merlo, Lauren M.F.; Pepper, John W.; Reid, Brian J.; Maley, Carlo C. (December 2006). "Cancer as an evolutionary and ecological process". Nature Reviews Cancer. 6 (12): 924–935. doi:10.1038/nrc2013. ISSN 1474-175X. PMID 17109012. S2CID 8040576.
- ^ Pan, Dabo; Weiwei Xue; Wenqi Zhang; et al. (October 2012). "Understanding the drug resistance mechanism of hepatitis C virus NS3/4A to ITMN-191 due to R155K, A156V, D168A/E mutations: a computational study". Biochimica et Biophysica Acta (BBA) - General Subjects. 1820 (10): 1526–1534. doi:10.1016/j.bbagen.2012.06.001. ISSN 0304-4165. PMID 22698669.
- ^ Woodford, Neil; Ellington, Matthew J. (January 2007). "The emergence of antibiotic resistance by mutation". Clinical Microbiology and Infection. 13 (1): 5–18. doi:10.1111/j.1469-0691.2006.01492.x. ISSN 1198-743X. PMID 17184282.
- ^ Labbé, Pierrick; Berticat, Claire; Berthomieu, Arnaud; et al. (16 November 2007). "Forty Years of Erratic Insecticide Resistance Evolution in the Mosquito Culex pipiens". PLOS Genetics. 3 (11): e205. doi:10.1371/journal.pgen.0030205. ISSN 1553-7390. PMC 2077897. PMID 18020711.
- ^ Neve, Paul (October 2007). "Challenges for herbicide resistance evolution and management: 50 years after Harper". Weed Research. 47 (5): 365–369. Bibcode:2007WeedR..47..365N. doi:10.1111/j.1365-3180.2007.00581.x. ISSN 0043-1737.
- ^ Rodríguez-Rojas, Alexandro; Rodríguez-Beltrán, Jerónimo; Couce, Alejandro; Blázquez, Jesús (August 2013). "Antibiotics and antibiotic resistance: A bitter fight against evolution". International Journal of Medical Microbiology. 303 (6–7): 293–297. doi:10.1016/j.ijmm.2013.02.004. ISSN 1438-4221. PMID 23517688.
- ^ Schenk, Martijn F.; Szendro, Ivan G.; Krug, Joachim; de Visser, J. Arjan G.M. (28 June 2012). "Quantifying the Adaptive Potential of an Antibiotic Resistance Enzyme". PLOS Genetics. 8 (6): e1002783. doi:10.1371/journal.pgen.1002783. ISSN 1553-7390. PMC 3386231. PMID 22761587.
- ^ Read, Andrew F.; Lynch, Penelope A.; Thomas, Matthew B. (7 April 2009). "How to Make Evolution-Proof Insecticides for Malaria Control". PLOS Biology. 7 (4): e1000058. doi:10.1371/journal.pbio.1000058. PMC 3279047. PMID 19355786.
- ^ Fraser, Alex S. (18 January 1958). "Monte Carlo Analyses of Genetic Models". Nature. 181 (4603): 208–209. Bibcode:1958Natur.181..208F. doi:10.1038/181208a0. ISSN 0028-0836. PMID 13504138. S2CID 4211563.
- ^ Rechenberg 1973
- ^ Holland 1975
- ^ Koza 1992
- ^ Jamshidi, Mo (15 August 2003). "Tools for intelligent control: fuzzy controllers, neural networks and genetic algorithms". Philosophical Transactions of the Royal Society A. 361 (1809): 1781–1808. Bibcode:2003RSPTA.361.1781J. doi:10.1098/rsta.2003.1225. PMID 12952685. S2CID 34259612.
- ^ "Age of the Earth". United States Geological Survey. 9 July 2007. Archived from the original on 23 December 2005. Retrieved 31 May 2015.
- ^ Dalrymple 2001, pp. 205–221
- ^ Manhesa, Gérard; Allègre, Claude J.; Dupréa, Bernard; Hamelin, Bruno (May 1980). "Lead isotope study of basic-ultrabasic layered complexes: Speculations about the age of the earth and primitive mantle characteristics". Earth and Planetary Science Letters. 47 (3): 370–382. Bibcode:1980E&PSL..47..370M. doi:10.1016/0012-821X(80)90024-2. ISSN 0012-821X.
- ^ Raven & Johnson 2002, p. 68
- ^ a b Borenstein, Seth (19 October 2015). "Hints of life on what was thought to be desolate early Earth". Excite. Yonkers, NY: Mindspark Interactive Network. Associated Press. Archived from the original on 23 October 2015. Retrieved 8 October 2018.
- ^ Bell, Elizabeth A.; Boehnike, Patrick; Harrison, T. Mark; Mao, Wendy L. (24 November 2015). "Potentially biogenic carbon preserved in a 4.1 billion-year-old zircon" (PDF). PNAS. 112 (47): 14518–14521. Bibcode:2015PNAS..11214518B. doi:10.1073/pnas.1517557112. ISSN 0027-8424. PMC 4664351. PMID 26483481. Archived (PDF) from the original on 6 November 2015. Retrieved 30 December 2015.
- ^ Schouten, Lucy (20 October 2015). "When did life first emerge on Earth? Maybe a lot earlier than we thought". The Christian Science Monitor. Boston, Massachusetts: Christian Science Publishing Society. ISSN 0882-7729. Archived from the original on 22 March 2016. Retrieved 11 July 2018.
- ^ Wade, Nicholas (25 July 2016). "Meet Luca, the Ancestor of All Living Things". The New York Times. New York. ISSN 0362-4331. Archived from the original on 28 July 2016. Retrieved 25 July 2016.
- ^ McKinney 1997, p. 110
- ^ Mora, Camilo; Tittensor, Derek P.; Adl, Sina; et al. (23 August 2011). "How Many Species Are There on Earth and in the Ocean?". PLOS Biology. 9 (8): e1001127. doi:10.1371/journal.pbio.1001127. ISSN 1545-7885. PMC 3160336. PMID 21886479.
- ^ Miller & Spoolman 2012, p. 62
- ^ Chapman 2009
- ^ Roskov, Y.; Abucay, L.; Orrell, T.; Nicolson, D.; et al., eds. (2016). "Species 2000 & ITIS Catalogue of Life, 2016 Annual Checklist". Species 2000. Leiden, Netherlands: Naturalis Biodiversity Center. ISSN 2405-884X. Archived from the original on 12 November 2016. Retrieved 6 November 2016.
- ^ Peretó, Juli (March 2005). "Controversies on the origin of life" (PDF). International Microbiology. 8 (1): 23–31. ISSN 1139-6709. PMID 15906258. Archived from the original (PDF) on 24 August 2015.
- ^ Marshall, Michael (11 November 2020). "Charles Darwin's hunch about early life was probably right – In a few scrawled notes to a friend, biologist Charles Darwin theorised how life began. Not only was it probably correct, his theory was a century ahead of its time". BBC News. Archived from the original on 11 November 2020. Retrieved 11 November 2020.
- ^ Joyce, Gerald F. (11 July 2002). "The antiquity of RNA-based evolution". Nature. 418 (6894): 214–221. Bibcode:2002Natur.418..214J. doi:10.1038/418214a. PMID 12110897. S2CID 4331004.
- ^ Trevors, Jack T.; Psenner, Roland (December 2001). "From self-assembly of life to present-day bacteria: a possible role for nanocells". FEMS Microbiology Reviews. 25 (5): 573–582. doi:10.1111/j.1574-6976.2001.tb00592.x. ISSN 1574-6976. PMID 11742692.
- ^ Theobald, Douglas L. (13 May 2010). "A formal test of the theory of universal common ancestry". Nature. 465 (7295): 219–222. Bibcode:2010Natur.465..219T. doi:10.1038/nature09014. ISSN 0028-0836. PMID 20463738. S2CID 4422345.
- ^ Bapteste, Eric; Walsh, David A. (June 2005). "Does the 'Ring of Life' ring true?". Trends in Microbiology. 13 (6): 256–261. doi:10.1016/j.tim.2005.03.012. ISSN 0966-842X. PMID 15936656.
- ^ Darwin 1859, p. 1
- ^ Doolittle, W. Ford; Bapteste, Eric (13 February 2007). "Pattern pluralism and the Tree of Life hypothesis". PNAS. 104 (7): 2043–2049. Bibcode:2007PNAS..104.2043D. doi:10.1073/pnas.0610699104. ISSN 0027-8424. PMC 1892968. PMID 17261804.
- ^ Kunin, Victor; Goldovsky, Leon; Darzentas, Nikos; Ouzounis, Christos A. (July 2005). "The net of life: Reconstructing the microbial phylogenetic network". Genome Research. 15 (7): 954–959. doi:10.1101/gr.3666505. ISSN 1088-9051. PMC 1172039. PMID 15965028.
- ^ Darwin 1837, p. 25
- ^ Jablonski, David (25 June 1999). "The Future of the Fossil Record". Science. 284 (5423): 2114–2116. doi:10.1126/science.284.5423.2114. ISSN 0036-8075. PMID 10381868. S2CID 43388925.
- ^ Mason, Stephen F. (6 September 1984). "Origins of biomolecular handedness". Nature. 311 (5981): 19–23. Bibcode:1984Natur.311...19M. doi:10.1038/311019a0. ISSN 0028-0836. PMID 6472461. S2CID 103653.
- ^ Wolf, Yuri I.; Rogozin, Igor B.; Grishin, Nick V.; Koonin, Eugene V. (1 September 2002). "Genome trees and the tree of life". Trends in Genetics. 18 (9): 472–479. doi:10.1016/S0168-9525(02)02744-0. ISSN 0168-9525. PMID 12175808.
- ^ Varki, Ajit; Altheide, Tasha K. (December 2005). "Comparing the human and chimpanzee genomes: searching for needles in a haystack". Genome Research. 15 (12): 1746–1758. CiteSeerX 10.1.1.673.9212. doi:10.1101/gr.3737405. ISSN 1088-9051. PMID 16339373.
- ^ Ciccarelli, Francesca D.; Doerks, Tobias; von Mering, Christian; et al. (3 March 2006). "Toward Automatic Reconstruction of a Highly Resolved Tree of Life" (PDF). Science. 311 (5765): 1283–1287. Bibcode:2006Sci...311.1283C. CiteSeerX 10.1.1.381.9514. doi:10.1126/science.1123061. ISSN 0036-8075. PMID 16513982. S2CID 1615592. Archived (PDF) from the original on 4 March 2016.
- ^ a b Cavalier-Smith, Thomas (29 June 2006). "Cell evolution and Earth history: stasis and revolution". Philosophical Transactions of the Royal Society B. 361 (1470): 969–1006. doi:10.1098/rstb.2006.1842. ISSN 0962-8436. PMC 1578732. PMID 16754610.
- ^ Schopf, J. William (29 June 2006). "Fossil evidence of Archaean life". Philosophical Transactions of the Royal Society B. 361 (1470): 869–885. doi:10.1098/rstb.2006.1834. PMC 1578735. PMID 16754604.
- Altermann, Wladyslaw; Kazmierczak, Józef (November 2003). "Archean microfossils: a reappraisal of early life on Earth". Research in Microbiology. 154 (9): 611–617. doi:10.1016/j.resmic.2003.08.006. PMID 14596897.
- ^ Schopf, J. William (19 July 1994). "Disparate rates, differing fates: tempo and mode of evolution changed from the Precambrian to the Phanerozoic". PNAS. 91 (15): 6735–6742. Bibcode:1994PNAS...91.6735S. doi:10.1073/pnas.91.15.6735. PMC 44277. PMID 8041691.
- ^ Poole, Anthony M.; Penny, David (January 2007). "Evaluating hypotheses for the origin of eukaryotes". BioEssays. 29 (1): 74–84. doi:10.1002/bies.20516. ISSN 0265-9247. PMID 17187354.
- ^ Dyall, Sabrina D.; Brown, Mark T.; Johnson, Patricia J. (9 April 2004). "Ancient Invasions: From Endosymbionts to Organelles". Science. 304 (5668): 253–257. Bibcode:2004Sci...304..253D. doi:10.1126/science.1094884. PMID 15073369. S2CID 19424594.
- ^ Martin, William (October 2005). "The missing link between hydrogenosomes and mitochondria". Trends in Microbiology. 13 (10): 457–459. doi:10.1016/j.tim.2005.08.005. PMID 16109488.
- ^ Lang, B. Franz; Gray, Michael W.; Burger, Gertraud (December 1999). "Mitochondrial genome evolution and the origin of eukaryotes". Annual Review of Genetics. 33: 351–397. doi:10.1146/annurev.genet.33.1.351. ISSN 0066-4197. PMID 10690412.
- McFadden, Geoffrey Ian (1 December 1999). "Endosymbiosis and evolution of the plant cell". Current Opinion in Plant Biology. 2 (6): 513–519. Bibcode:1999COPB....2..513M. doi:10.1016/S1369-5266(99)00025-4. PMID 10607659.
- ^ DeLong, Edward F.; Pace, Norman R. (1 August 2001). "Environmental Diversity of Bacteria and Archaea". Systematic Biology. 50 (4): 470–478. CiteSeerX 10.1.1.321.8828. doi:10.1080/106351501750435040. ISSN 1063-5157. PMID 12116647.
- ^ Kaiser, Dale (December 2001). "Building a multicellular organism". Annual Review of Genetics. 35: 103–123. doi:10.1146/annurev.genet.35.102401.090145. ISSN 0066-4197. PMID 11700279. S2CID 18276422.
- ^ Zimmer, Carl (7 January 2016). "Genetic Flip Helped Organisms Go From One Cell to Many". The New York Times. New York. ISSN 0362-4331. Archived from the original on 7 January 2016. Retrieved 7 January 2016.
- ^ Valentine, James W.; Jablonski, David; Erwin, Douglas H. (1 March 1999). "Fossils, molecules and embryos: new perspectives on the Cambrian explosion". Development. 126 (5): 851–859. doi:10.1242/dev.126.5.851. ISSN 0950-1991. PMID 9927587. Archived from the original on 1 March 2015. Retrieved 30 December 2014.
- ^ Ohno, Susumu (January 1997). "The reason for as well as the consequence of the Cambrian explosion in animal evolution". Journal of Molecular Evolution. 44 (Suppl. 1): S23–S27. Bibcode:1997JMolE..44S..23O. doi:10.1007/PL00000055. ISSN 0022-2844. PMID 9071008. S2CID 21879320.
- Valentine, James W.; Jablonski, David (2003). "Morphological and developmental macroevolution: a paleontological perspective". The International Journal of Developmental Biology. 47 (7–8): 517–522. ISSN 0214-6282. PMID 14756327. Archived from the original on 24 October 2014. Retrieved 30 December 2014.
- ^ Waters, Elizabeth R. (December 2003). "Molecular adaptation and the origin of land plants". Molecular Phylogenetics and Evolution. 29 (3): 456–463. Bibcode:2003MolPE..29..456W. doi:10.1016/j.ympev.2003.07.018. ISSN 1055-7903. PMID 14615186.
- ^ Mayhew, Peter J. (August 2007). "Why are there so many insect species? Perspectives from fossils and phylogenies". Biological Reviews. 82 (3): 425–454. doi:10.1111/j.1469-185X.2007.00018.x. ISSN 1464-7931. PMID 17624962. S2CID 9356614.
- ^ Carroll, Robert L. (May 2007). "The Palaeozoic Ancestry of Salamanders, Frogs and Caecilians". Zoological Journal of the Linnean Society. 150 (Supplement s1): 1–140. doi:10.1111/j.1096-3642.2007.00246.x. ISSN 1096-3642.
- ^ Wible, John R.; Rougier, Guillermo W.; Novacek, Michael J.; Asher, Robert J. (21 June 2007). "Cretaceous eutherians and Laurasian origin for placental mammals near the K/T boundary". Nature. 447 (7147): 1003–1006. Bibcode:2007Natur.447.1003W. doi:10.1038/nature05854. ISSN 0028-0836. PMID 17581585. S2CID 4334424.
- ^ Witmer, Lawrence M. (28 July 2011). "Palaeontology: An icon knocked from its perch". Nature. 475 (7357): 458–459. doi:10.1038/475458a. ISSN 0028-0836. PMID 21796198. S2CID 205066360.
- ^ Darwin 1909, p. 53
- ^ Kirk, Raven & Schofield 1983, pp. 100–142, 280–321
- ^ Lucretius
- ^ Sedley, David (2003). "Lucretius and the New Empedocles" (PDF). Leeds International Classical Studies. 2 (4). ISSN 1477-3643. Archived from the original (PDF) on 23 August 2014. Retrieved 25 November 2014.
- ^ Torrey, Harry Beal; Felin, Frances (March 1937). "Was Aristotle an Evolutionist?". The Quarterly Review of Biology. 12 (1): 1–18. doi:10.1086/394520. ISSN 0033-5770. JSTOR 2808399. S2CID 170831302.
- ^ Hull, David L. (December 1967). "The Metaphysics of Evolution". The British Journal for the History of Science. 3 (4). Cambridge: Cambridge University Press on behalf of The British Society for the History of Science: 309–337. doi:10.1017/S0007087400002892. JSTOR 4024958. S2CID 170328394.
- ^ Mason 1962, pp. 43–44
- ^ Kiros, Teodros. Explorations in African Political Thought. 2001, page 55
- ^ Mayr 1982, pp. 256–257
- ^ Waggoner, Ben (7 July 2000). "Carl Linnaeus (1707–1778)". Evolution (Online exhibit). Berkeley, California: University of California Museum of Paleontology. Archived from the original on 30 April 2011. Retrieved 11 February 2012.
- ^ Bowler 2003, pp. 73–75
- ^ "Erasmus Darwin (1731–1802)". Evolution (Online exhibit). Berkeley, California: University of California Museum of Paleontology. 4 October 1995. Archived from the original on 19 January 2012. Retrieved 11 February 2012.
- ^ Lamarck 1809
- ^ a b Nardon & Grenier 1991, p. 162
- ^ Ghiselin, Michael T. (September–October 1994). "The Imaginary Lamarck: A Look at Bogus 'History' in Schoolbooks". The Textbook Letter. OCLC 23228649. Archived from the original on 12 February 2008. Retrieved 23 January 2008.
{{cite journal}}: CS1 maint: unfit URL (link) - ^ Jablonka, Eva; Lamb, Marion J. (August 2007). "Précis of Evolution in Four Dimensions". Behavioral and Brain Sciences. 30 (4): 353–365. doi:10.1017/S0140525X07002221. ISSN 0140-525X. PMID 18081952. S2CID 15879804.
- ^ Burkhardt & Smith 1991
- "Darwin, C. R. to Lubbock, John". Darwin Correspondence Project. Cambridge: University of Cambridge. Archived from the original on 15 December 2014. Retrieved 1 December 2014. Letter 2532, 22 November 1859.
- ^ Sulloway, Frank J. (June 2009). "Why Darwin rejected intelligent design". Journal of Biosciences. 34 (2): 173–183. doi:10.1007/s12038-009-0020-8. ISSN 0250-5991. PMID 19550032. S2CID 12289290.
- ^ "Search results for "descent with modification" – The Complete Work of Charles Darwin Online". Archived from the original on 5 June 2022. Retrieved 30 July 2022.
- ^ Sober, Elliott (16 June 2009). "Did Darwin write the Origin backwards?". PNAS. 106 (Suppl. 1): 10048–10055. Bibcode:2009PNAS..10610048S. doi:10.1073/pnas.0901109106. ISSN 0027-8424. PMC 2702806. PMID 19528655.
- ^ Mayr 2002, p. 165
- ^ Bowler 2003, pp. 145–146
- ^ Sokal, Robert R.; Crovello, Theodore J. (March–April 1970). "The Biological Species Concept: A Critical Evaluation". The American Naturalist. 104 (936): 127–153. Bibcode:1970ANat..104..127S. doi:10.1086/282646. ISSN 0003-0147. JSTOR 2459191. S2CID 83528114.
- ^ Darwin, Charles; Wallace, Alfred (20 August 1858). "On the Tendency of Species to form Varieties; and on the Perpetuation of Varieties and Species by Natural Means of Selection". Journal of the Proceedings of the Linnean Society of London. Zoology. 3 (9): 45–62. doi:10.1111/j.1096-3642.1858.tb02500.x. ISSN 1096-3642. Archived from the original on 14 July 2007. Retrieved 13 May 2007.
- ^ Desmond, Adrian J. (17 July 2014). "Thomas Henry Huxley". Encyclopædia Britannica Online. Chicago, Illinois: Encyclopædia Britannica, Inc. Archived from the original on 19 January 2015. Retrieved 2 December 2014.
- ^ Plate, Robert. The Dinosaur Hunters: Othniel C. Marsh and Edward D. Cope, pp. 69, 203–5, David McKay Company, Inc., New York, 1964.
- ^ McCarren, Mark J. The Scientific Contributions of Othniel Charles Marsh, pp. 37–9, Peabody Museum of Natural History, Yale University, New Haven, Connecticut, 1993. ISBN 0-912532-32-7
- ^ Plate, Robert. The Dinosaur Hunters: Othniel C. Marsh and Edward D. Cope, pp. 188–9, David McKay Company, Inc., New York, 1964.
- ^ Y. -S. Liu; X. M. Zhou; M. X. Zhi; X. J. Li; Q. L. Wang (September 2009). "Darwin's contributions to genetics". Journal of Applied Genetics. 50 (3): 177–184. doi:10.1007/BF03195671. ISSN 1234-1983. PMID 19638672. S2CID 19919317.
- ^ Weiling, Franz (July 1991). "Historical study: Johann Gregor Mendel 1822–1884". American Journal of Medical Genetics. 40 (1): 1–25, discussion 26. doi:10.1002/ajmg.1320400103. PMID 1887835.
- ^ Wright 1984, p. 480
- ^ Provine 1971
- ^ Stamhuis, Ida H.; Meijer, Onno G.; Zevenhuizen, Erik J. A. (June 1999). "Hugo de Vries on Heredity, 1889–1903: Statistics, Mendelian Laws, Pangenes, Mutations". Isis. 90 (2): 238–267. doi:10.1086/384323. JSTOR 237050. PMID 10439561. S2CID 20200394.
- ^ a b Bowler 1989, pp. 307–318.
- ^ Levinson 2019.
- ^ Watson, J. D.; Crick, F. H. C. (25 April 1953). "Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid" (PDF). Nature. 171 (4356): 737–738. Bibcode:1953Natur.171..737W. doi:10.1038/171737a0. ISSN 0028-0836. PMID 13054692. S2CID 4253007. Archived (PDF) from the original on 23 August 2014. Retrieved 4 December 2014.
It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material.
- ^ Hennig 1999, p. 280
- ^ Dobzhansky, Theodosius (March 1973). "Nothing in Biology Makes Sense Except in the Light of Evolution" (PDF). The American Biology Teacher. 35 (3): 125–129. CiteSeerX 10.1.1.324.2891. doi:10.2307/4444260. JSTOR 4444260. S2CID 207358177. Archived from the original (PDF) on 23 October 2015.
- ^ Avise, John C.; Ayala, Francisco J. (11 May 2010). "In the light of evolution IV: The human condition" (PDF). PNAS. 107 (Suppl. 2): 8897–8901. doi:10.1073/pnas.1003214107. ISSN 0027-8424. PMC 3024015. PMID 20460311. Archived (PDF) from the original on 23 August 2014. Retrieved 29 December 2014.
- ^ Danchin, Étienne; Charmantier, Anne; Champagne, Frances A.; Mesoudi, Alex; Pujol, Benoit; Blanchet, Simon (June 2011). "Beyond DNA: integrating inclusive inheritance into an extended theory of evolution". Nature Reviews Genetics. 12 (7): 475–486. doi:10.1038/nrg3028. ISSN 1471-0056. PMID 21681209. S2CID 8837202.
- ^ Pigliucci & Müller 2010
- ^ Browne 2003, pp. 376–379
- ^ For an overview of the philosophical, religious and cosmological controversies, see:
For the scientific and social reception of evolution in the 19th and early 20th centuries, see:
- Johnston, Ian C. (1999). "Section Three: The Origins of Evolutionary Theory". ... And Still We Evolve: A Handbook for the Early History of Modern Science (3rd revised ed.). Nanaimo, BC: Liberal Studies Department, Malaspina University-College. Archived from the original on 16 April 2016. Retrieved 1 January 2015.
- Bowler 2003
- Zuckerkandl, Emile (30 December 2006). "Intelligent design and biological complexity". Gene. 385: 2–18. doi:10.1016/j.gene.2006.03.025. ISSN 0378-1119. PMID 17011142.
- ^ Ross, Marcus R. (May 2005). "Who Believes What? Clearing up Confusion over Intelligent Design and Young-Earth Creationism" (PDF). Journal of Geoscience Education. 53 (3): 319–323. Bibcode:2005JGeEd..53..319R. CiteSeerX 10.1.1.404.1340. doi:10.5408/1089-9995-53.3.319. ISSN 1089-9995. S2CID 14208021. Archived (PDF) from the original on 11 May 2008. Retrieved 28 April 2008.
- ^ Hameed, Salman (12 December 2008). "Bracing for Islamic Creationism" (PDF). Science. 322 (5908): 1637–1638. doi:10.1126/science.1163672. ISSN 0036-8075. PMID 19074331. S2CID 206515329. Archived from the original (PDF) on 10 November 2014.
- ^ Bowler 2003
- ^ Miller, Jon D.; Scott, Eugenie C.; Okamoto, Shinji (11 August 2006). "Public Acceptance of Evolution". Science. 313 (5788): 765–766. doi:10.1126/science.1126746. ISSN 0036-8075. PMID 16902112. S2CID 152990938.
- ^ Spergel, David Nathaniel; Verde, Licia; Peiris, Hiranya V.; et al. (2003). "First-Year Wilkinson Microwave Anisotropy Probe (WMAP) Observations: Determination of Cosmological Parameters". The Astrophysical Journal Supplement Series. 148 (1): 175–194. arXiv:astro-ph/0302209. Bibcode:2003ApJS..148..175S. doi:10.1086/377226. S2CID 10794058.
- ^ Wilde, Simon A.; Valley, John W.; Peck, William H.; Graham, Colin M. (11 January 2001). "Evidence from detrital zircons for the existence of continental crust and oceans on the Earth 4.4 Gyr ago". Nature. 409 (6817): 175–178. Bibcode:2001Natur.409..175W. doi:10.1038/35051550. ISSN 0028-0836. PMID 11196637. S2CID 4319774.
- ^ Branch, Glenn (March 2007). "Understanding Creationism after Kitzmiller". BioScience. 57 (3): 278–284. doi:10.1641/B570313. ISSN 0006-3568. S2CID 86665329.
- ^ Xiaoxing Jin (March 2019). "Translation and transmutation: the Origin of Species in China". The British Journal for the History of Science. 52 (1). Cambridge: Cambridge University Press on behalf of The British Society for the History of Science: 117–141. doi:10.1017/S0007087418000808. PMID 30587253. S2CID 58605626.
Bibliography
- Altenberg, Lee (1995). "Genome growth and the evolution of the genotype–phenotype map". In Banzhaf, Wolfgang; Eeckman, Frank H. (eds.). Evolution and Biocomputation: Computational Models of Evolution. Lecture Notes in Computer Science. Vol. 899. Berlin; New York: Springer-Verlag Berlin Heidelberg. pp. 205–259. CiteSeerX 10.1.1.493.6534. doi:10.1007/3-540-59046-3_11. ISBN 978-3-540-59046-0. ISSN 0302-9743. LCCN 95005970. OCLC 32049812.
- Birdsell, John A.; Wills, Christopher (2003). "The Evolutionary Origin and Maintenance of Sexual Recombination: A Review of Contemporary Models". In MacIntyre, Ross J.; Clegg, Michael T. (eds.). Evolutionary Biology. Vol. 33. New York: Springer Science+Business Media. ISBN 978-1-4419-3385-0. ISSN 0071-3260. OCLC 751583918.
- Bowler, Peter J. (1989). The Mendelian Revolution: The Emergence of Hereditarian Concepts in Modern Science and Society. Baltimore, Maryland: Johns Hopkins University Press. ISBN 978-0-8018-3888-0. LCCN 89030914. OCLC 19322402.
- Bowler, Peter J. (2003). Evolution: The History of an Idea (3rd completely rev. and expanded ed.). Berkeley, California: University of California Press. ISBN 978-0-520-23693-6. LCCN 2002007569. OCLC 49824702.
- Browne, Janet (2003). Charles Darwin: The Power of Place. Vol. 2. London: Pimlico. ISBN 978-0-7126-6837-8. LCCN 94006598. OCLC 52327000.
- Burkhardt, Frederick; Smith, Sydney, eds. (1991). The Correspondence of Charles Darwin. Vol. 7: 1858–1859. Cambridge: Cambridge University Press. ISBN 978-0-521-38564-0. LCCN 84045347. OCLC 185662993.
- Carroll, Sean B.; Grenier, Jennifer K.; Weatherbee, Scott D. (2005). From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design (2nd ed.). Malden, Massachusetts: Blackwell Publishing. ISBN 978-1-4051-1950-4. LCCN 2003027991. OCLC 53972564.
- Chapman, Arthur D. (2009). Numbers of Living Species in Australia and the World (2nd ed.). Canberra: Department of the Environment, Water, Heritage and the Arts: Australian Biological Resources Study. ISBN 978-0-642-56860-1. OCLC 780539206. Archived from the original on 25 December 2016. Retrieved 6 November 2016.
- Coyne, Jerry A. (2009). Why Evolution is True. New York: Viking. ISBN 978-0-670-02053-9. LCCN 2008033973. OCLC 233549529.
- Dalrymple, G. Brent (2001). "The age of the Earth in the twentieth century: a problem (mostly) solved". In Lewis, C.L.E.; Knell, S.J. (eds.). The Age of the Earth: from 4004 BC to AD 2002. Geological Society Special Publication. Vol. 190. pp. 205–221. Bibcode:2001GSLSP.190..205D. doi:10.1144/gsl.sp.2001.190.01.14. ISBN 978-1-86239-093-5. LCCN 2003464816. OCLC 48570033. S2CID 130092094.
- Darwin, Charles (1837). "B" Notebook. The notebook is available from The Complete Work of Charles Darwin Online Archived 18 March 2022 at the Wayback Machine. Retrieved 2019-10-09.
- Darwin, Charles (1859). On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life (1st ed.). London: John Murray. LCCN 06017473. OCLC 741260650. The book is available from The Complete Work of Charles Darwin Online Archived 27 January 2015 at the Wayback Machine. Retrieved 2014-11-21.
- Darwin, Charles (1872). The Expression of the Emotions in Man and Animals. London: John Murray. LCCN 04002793. OCLC 1102785.
- Darwin, Francis, ed. (1909). The foundations of The origin of species, a sketch written in 1842 (PDF). Cambridge: Printed at the University Press. LCCN 61057537. OCLC 1184581. Archived (PDF) from the original on 4 March 2016. Retrieved 27 November 2014.
- Dennett, Daniel (1995). Darwin's Dangerous Idea: Evolution and the Meanings of Life. New York: Simon & Schuster. ISBN 978-0-684-80290-9. LCCN 94049158. OCLC 31867409.
- Dobzhansky, Theodosius (1968). "On Some Fundamental Concepts of Darwinian Biology". In Dobzhansky, Theodosius; Hecht, Max K.; Steere, William C. (eds.). Evolutionary Biology (1st ed.). New York: Appleton-Century-Crofts. pp. 1–34. doi:10.1007/978-1-4684-8094-8_1. ISBN 978-1-4684-8096-2. OCLC 24875357.
- Dobzhansky, Theodosius (1970). Genetics of the Evolutionary Process. New York: Columbia University Press. ISBN 978-0-231-02837-0. LCCN 72127363. OCLC 97663.
- Eldredge, Niles; Gould, Stephen Jay (1972). "Punctuated equilibria: an alternative to phyletic gradualism". In Schopf, Thomas J.M. (ed.). Models in Paleobiology. San Francisco, California: Freeman, Cooper. ISBN 978-0-87735-325-6. LCCN 72078387. OCLC 572084.
- Eldredge, Niles (1985). Time Frames: The Rethinking of Darwinian Evolution and the Theory of Punctuated Equilibria. New York: Simon & Schuster. ISBN 978-0-671-49555-8. LCCN 84023632. OCLC 11443805.
- Ewens, Warren J. (2004). Mathematical Population Genetics. Interdisciplinary Applied Mathematics. Vol. I. Theoretical Introduction (2nd ed.). New York: Springer-Verlag New York. ISBN 978-0-387-20191-7. LCCN 2003065728. OCLC 53231891.
- Fisher, Ronald A. (1930). The Genetical Theory of Natural Selection. Oxford: The Clarendon Press. ISBN 978-0-19-850440-5. LCCN 30029177. OCLC 18500548.
- Futuyma, Douglas J. (2004). "The Fruit of the Tree of Life: Insights into Evolution and Ecology". In Cracraft, Joel; Donoghue, Michael J. (eds.). Assembling the Tree of Life. Oxford; New York: Oxford University Press. ISBN 978-0-19-517234-8. LCCN 2003058012. OCLC 61342697. "Proceedings of a symposium held at the American Museum of Natural History in New York, 2002."
- Futuyma, Douglas J. (2005). Evolution. Sunderland, Massachusetts: Sinauer Associates. ISBN 978-0-87893-187-3. LCCN 2004029808. OCLC 57311264.
- Futuyma, Douglas J.; Kirkpatrick, Mark (2017). Evolution (Fourth ed.). Sunderland, Massachusetts: Sinauer Associates. ISBN 978-1-60535-605-1. LCCN 2017000562. OCLC 969439375.
- Gould, Stephen Jay (2002). The Structure of Evolutionary Theory. Cambridge, Massachusetts: Belknap Press of Harvard University Press. ISBN 978-0-674-00613-3. LCCN 2001043556. OCLC 47869352.
- Gray, Peter (2007). Psychology (5th ed.). New York: Worth Publishers. ISBN 978-0-7167-0617-5. LCCN 2006921149. OCLC 76872504.
- Hall, Brian K.; Hallgrímsson, Benedikt (2008). Strickberger's Evolution (4th ed.). Sudbury, Massachusetts: Jones and Bartlett Publishers. ISBN 978-0-7637-0066-9. LCCN 2007008981. OCLC 85814089.
- Hennig, Willi (1999) [Originally published 1966 (reprinted 1979); translated from the author's unpublished revision of Grundzüge einer Theorie der phylogenetischen Systematik, published in 1950]. Phylogenetic Systematics. Translation by D. Dwight Davis and Rainer Zangerl; foreword by Donn E. Rosen, Gareth Nelson, and Colin Patterson (Reissue ed.). Urbana, Illinois: University of Illinois Press. ISBN 978-0-252-06814-0. LCCN 78031969. OCLC 722701473.
- Holland, John H. (1975). Adaptation in Natural and Artificial Systems: An Introductory Analysis with Applications to Biology, Control, and Artificial Intelligence. Ann Arbor, Michigan: University of Michigan Press. ISBN 978-0-472-08460-9. LCCN 74078988. OCLC 1531617.
- Kampourakis, Kostas (2014). Understanding Evolution. Cambridge; New York: Cambridge University Press. ISBN 978-1-107-03491-4. LCCN 2013034917. OCLC 855585457.
- Kirk, Geoffrey; Raven, John; Schofield, Malcolm (1983). The Presocratic Philosophers: A Critical History with a Selection of Texts (2nd ed.). Cambridge; New York: Cambridge University Press. ISBN 978-0-521-27455-5. LCCN 82023505. OCLC 9081712.
- Koza, John R. (1992). Genetic Programming: On the Programming of Computers by Means of Natural Selection. Complex Adaptive Systems. Cambridge, Massachusetts: MIT Press. ISBN 978-0-262-11170-6. LCCN 92025785. OCLC 26263956.
- Lamarck, Jean-Baptiste (1809). Philosophie Zoologique. Paris: Dentu et L'Auteur. OCLC 2210044. Philosophie zoologique (1809) at the Internet Archive. Retrieved 2014-11-29.
- Lane, David H. (1996). The Phenomenon of Teilhard: Prophet for a New Age (1st ed.). Macon, Georgia: Mercer University Press. ISBN 978-0-86554-498-7. LCCN 96008777. OCLC 34710780.
- Levinson, Gene (2019). Rethinking Evolution: The Revolution That's Hiding in Plain Sight. Hackensack, New Jersey: World Scientific. ISBN 978-1-78634-726-8. LCCN 2019013762. OCLC 1138095098. Archived from the original on 21 May 2022. Retrieved 30 July 2022.
- Lucretius. "Book V, lines 855–877". De Rerum Natura. Edited and translated by William Ellery Leonard (1916). Medford/Somerville, Massachusetts: Tufts University. OCLC 33233743. Archived from the original on 4 September 2014. Retrieved 25 November 2014 – via Perseus Digital Library.
- Mason, Stephen F. (1962). A History of the Sciences. Collier Books. Science Library, CS9 (New rev. ed.). New York: Collier Books. LCCN 62003378. OCLC 568032626.
- Maynard Smith, John (1978). The Evolution of Sex. Cambridge; New York: Cambridge University Press. ISBN 978-0-521-29302-0. LCCN 77085689. OCLC 3413793.
- Maynard Smith, John (1998). "The Units of Selection". In Bock, Gregory R.; Goode, Jamie A. (eds.). The Limits of Reductionism in Biology. Novartis Foundation Symposia. Vol. 213. Chichester; New York: John Wiley & Sons. pp. 203–221. doi:10.1002/9780470515488.ch15. ISBN 978-0-471-97770-4. LCCN 98002779. OCLC 38311600. PMID 9653725. "Papers from the Symposium on the Limits of Reductionism in Biology, held at the Novartis Foundation, London, May 13–15, 1997."
- Mayr, Ernst (1942). Systematics and the Origin of Species from the Viewpoint of a Zoologist. Columbia Biological Series. Vol. 13. New York: Columbia University Press. LCCN 43001098. OCLC 766053.
- Mayr, Ernst (1982). The Growth of Biological Thought: Diversity, Evolution, and Inheritance. Translation of John Ray by E. Silk. Cambridge, Massachusetts: Belknap Press. ISBN 978-0-674-36445-5. LCCN 81013204. OCLC 7875904.
- Mayr, Ernst (2002) [Originally published 2001; New York: Basic Books]. What Evolution Is. Science Masters. London: Weidenfeld & Nicolson. ISBN 978-0-297-60741-0. LCCN 2001036562. OCLC 248107061.
- McKinney, Michael L. (1997). "How do rare species avoid extinction? A paleontological view". In Kunin, William E.; Gaston, Kevin J. (eds.). The Biology of Rarity: Causes and consequences of rare—common differences (1st ed.). London; New York: Chapman & Hall. ISBN 978-0-412-63380-5. LCCN 96071014. OCLC 36442106.
- Miller, G. Tyler; Spoolman, Scott E. (2012). Environmental Science (14th ed.). Belmont, California: Brooks/Cole. ISBN 978-1-111-98893-7. LCCN 2011934330. OCLC 741539226. Archived from the original on 2 May 2019. Retrieved 27 December 2014.
- Nardon, Paul; Grenier, Anne-Marie (1991). "Serial Endosymbiosis Theory and Weevil Evolution: The Role of Symbiosis". In Margulis, Lynn; Fester, René (eds.). Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis. Cambridge, Massachusetts: MIT Press. ISBN 978-0-262-13269-5. LCCN 90020439. OCLC 22597587. "Based on a conference held in Bellagio, Italy, June 25–30, 1989"
- National Academy of Sciences; Institute of Medicine (2008). Science, Evolution, and Creationism. Washington, DC: National Academy Press. ISBN 978-0-309-10586-6. LCCN 2007015904. OCLC 123539346. Retrieved 22 November 2014.
- Odum, Eugene P. (1971). Fundamentals of Ecology (3rd ed.). Philadelphia, Pennsylvania: Saunders. ISBN 978-0-7216-6941-0. LCCN 76081826. OCLC 154846.
- Okasha, Samir (2006). Evolution and the Levels of Selection. Oxford; New York: Oxford University Press. ISBN 978-0-19-926797-2. LCCN 2006039679. OCLC 70985413.
- Panno, Joseph (2005). The Cell: Evolution of the First Organism. Facts on File science library. New York: Facts on File. ISBN 978-0-8160-4946-2. LCCN 2003025841. OCLC 53901436.
- Piatigorsky, Joram; Kantorow, Marc; Gopal-Srivastava, Rashmi; Tomarev, Stanislav I. (1994). "Recruitment of enzymes and stress proteins as lens crystallins". In Jansson, Bengt; Jörnvall, Hans; Rydberg, Ulf; et al. (eds.). Toward a Molecular Basis of Alcohol Use and Abuse. Experientia. Vol. 71. Basel; Boston: Birkhäuser Verlag. pp. 241–50. doi:10.1007/978-3-0348-7330-7_24. ISBN 978-3-7643-2940-2. LCCN 94010167. OCLC 30030941. PMID 8032155.
- Pigliucci, Massimo; Müller, Gerd B., eds. (2010). Evolution, the Extended Synthesis. Cambridge, Massachusetts: MIT Press. ISBN 978-0-262-51367-8. LCCN 2009024587. OCLC 804875316. Archived from the original on 18 September 2015.
- Provine, William B. (1971). The Origins of Theoretical Population Genetics. Chicago History of Science and Medicine (2nd ed.). Chicago, Illinois: University of Chicago Press. ISBN 978-0-226-68464-2. LCCN 2001027561. OCLC 46660910.
- Raven, Peter H.; Johnson, George B. (2002). Biology (6th ed.). Boston, Massachusetts: McGraw-Hill. ISBN 978-0-07-112261-0. LCCN 2001030052. OCLC 45806501.
- Ray, John (1686). Historia Plantarum [History of Plants]. Vol. I. Londini: Typis Mariæ Clark. LCCN agr11000774. OCLC 2126030.
- Rechenberg, Ingo (1973). Evolutionsstrategie; Optimierung technischer Systeme nach Prinzipien der biologischen Evolution (PhD thesis). Problemata (in German). Vol. 15. Afterword by Manfred Eigen. Stuttgart-Bad Cannstatt: Frommann-Holzboog. ISBN 978-3-7728-0373-4. LCCN 74320689. OCLC 9020616.
- Ridley, Mark (2004). Evolution. Oxford: Blackwell. ISBN 978-1-4051-0345-9.
- Stearns, Beverly Peterson; Stearns, Stephen C. (1999). Watching, from the Edge of Extinction. New Haven, Connecticut: Yale University Press. ISBN 978-0-300-08469-6. LCCN 98034087. OCLC 803522914.
- Stevens, Anthony (1982). Archetype: A Natural History of the Self. London: Routledge & Kegan Paul. ISBN 978-0-7100-0980-7. LCCN 84672250. OCLC 10458367.
- Voet, Donald; Voet, Judith G.; Pratt, Charlotte W. (2016). Fundamentals of Biochemistry: Life at the Molecular Level (Fifth ed.). Hoboken, New Jersey: John Wiley & Sons. ISBN 978-1-118-91840-1. LCCN 2016002847. OCLC 939245154.
- Wright, Sewall (1984). Genetic and Biometric Foundations. Evolution and the Genetics of Populations. Vol. 1. Chicago, Illinois: University of Chicago Press. ISBN 978-0-226-91038-3. LCCN 67025533. OCLC 246124737.
Further reading
- Introductory reading
- Barrett, Paul H.; Weinshank, Donald J.; Gottleber, Timothy T., eds. (1981). A Concordance to Darwin's Origin of Species, First Edition. Ithaca, New York: Cornell University Press. ISBN 978-0-8014-1319-3. LCCN 80066893. OCLC 610057960.
- Carroll, Sean B. (2005). Endless Forms Most Beautiful: The New Science of Evo Devo and the Making of the Animal Kingdom. illustrations by Jamie W. Carroll, Josh P. Klaiss, Leanne M. Olds (1st ed.). New York: W.W. Norton & Company. ISBN 978-0-393-06016-4. LCCN 2004029388. OCLC 57316841.
- Charlesworth, Brian; Charlesworth, Deborah (2003). Evolution: A Very Short Introduction. Very Short Introductions. Oxford; New York: Oxford University Press. ISBN 978-0-19-280251-4. LCCN 2003272247. OCLC 51668497.
- Gould, Stephen Jay (1989). Wonderful Life: The Burgess Shale and the Nature of History (1st ed.). New York: W.W. Norton & Company. ISBN 978-0-393-02705-1. LCCN 88037469. OCLC 18983518.
- Jones, Steve (1999). Almost Like a Whale: The Origin of Species Updated. London; New York: Doubleday. ISBN 978-0-385-40985-8. LCCN 2002391059. OCLC 41420544.
- —— (2000). Darwin's Ghost: The Origin of Species Updated (1st ed.). New York: Random House. ISBN 978-0-375-50103-6. LCCN 99053246. OCLC 42690131. American version.
- Mader, Sylvia S. (2007). Biology. Significant contributions by Murray P. Pendarvis (9th ed.). Boston, Massachusetts: McGraw-Hill Higher Education. ISBN 978-0-07-246463-4. LCCN 2005027781. OCLC 61748307.
- Maynard Smith, John (1993). The Theory of Evolution (Canto ed.). Cambridge; New York: Cambridge University Press. ISBN 978-0-521-45128-4. LCCN 93020358. OCLC 27676642.
- Pallen, Mark J. (2009). The Rough Guide to Evolution. Rough Guides Reference Guides. London; New York: Rough Guides. ISBN 978-1-85828-946-5. LCCN 2009288090. OCLC 233547316.
- Advanced reading
- Barton, Nicholas H.; Briggs, Derek E.G.; Eisen, Jonathan A.; et al. (2007). Evolution. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press. ISBN 978-0-87969-684-9. LCCN 2007010767. OCLC 86090399.
- Coyne, Jerry A.; Orr, H. Allen (2004). Speciation. Sunderland, Massachusetts: Sinauer Associates. ISBN 978-0-87893-089-0. LCCN 2004009505. OCLC 55078441.
- Bergstrom, Carl T.; Dugatkin, Lee Alan (2012). Evolution (1st ed.). New York: W.W. Norton & Company. ISBN 978-0-393-91341-5. LCCN 2011036572. OCLC 729341924.
- Hall, Brian K.; Olson, Wendy, eds. (2003). Keywords and Concepts in Evolutionary Developmental Biology. Cambridge, Massachusetts: Harvard University Press. ISBN 978-0-674-00904-2. LCCN 2002192201. OCLC 50761342.
- Kauffman, Stuart A. (1993). The Origins of Order: Self-organization and Selection in Evolution. New York; Oxford: Oxford University Press. ISBN 978-0-19-507951-7. LCCN 91011148. OCLC 895048122.
- Maynard Smith, John; Szathmáry, Eörs (1995). The Major Transitions in Evolution. Oxford; New York: W.H. Freeman Spektrum. ISBN 978-0-7167-4525-9. LCCN 94026965. OCLC 30894392.
- Mayr, Ernst (2001). What Evolution Is. New York: Basic Books. ISBN 978-0-465-04426-9. LCCN 2001036562. OCLC 47443814.
- Minelli, Alessandro (2009). Forms of Becoming: The Evolutionary Biology of Development. Translation by Mark Epstein. Princeton, New Jersey; Oxford: Princeton University Press. ISBN 978-0-691-13568-7. LCCN 2008028825. OCLC 233030259.
External links
- General information
- "Evolution" on In Our Time at the BBC
- "Evolution Resources from the National Academies". Washington, DC: National Academy of Sciences. Retrieved 30 May 2011.
- "Understanding Evolution: your one-stop resource for information on evolution". Berkeley, California: University of California, Berkeley. Retrieved 30 May 2011.
- "Evolution of Evolution – 150 Years of Darwin's 'On the Origin of Species'". Arlington County, Virginia: National Science Foundation. Archived from the original on 19 May 2011. Retrieved 30 May 2011.
- "Human Evolution Timeline Interactive". Smithsonian Institution, National Museum of Natural History. 28 January 2010. Retrieved 14 July 2018. Adobe Flash required.
- "History of Evolution in the United States". Salon. Retrieved 2021-08-24.
- Video (1980; Cosmos animation; 8:01): "Evolution" – Carl Sagan on YouTube
- Experiments
- Lenski, Richard E. "Experimental Evolution". East Lansing, Michigan: Michigan State University. Retrieved 31 July 2013.
- Chastain, Erick; Livnat, Adi; Papadimitriou, Christos; Vazirani, Umesh (22 July 2014). "Algorithms, games, and evolution". PNAS. 111 (29): 10620–10623. Bibcode:2014PNAS..11110620C. doi:10.1073/pnas.1406556111. ISSN 0027-8424. PMC 4115542. PMID 24979793.
- Online lectures
- "Evolution Matters Lecture Series". Harvard Online Learning. Cambridge, Massachusetts: Harvard University. Archived from the original on 18 December 2017. Retrieved 15 July 2018.
- Stearns, Stephen C. "EEB 122: Principles of Evolution, Ecology and Behavior". Open Yale Courses. New Haven, Connecticut: Yale University. Archived from the original on 1 December 2017. Retrieved 14 July 2018.


