Nucleic acid structure determination
Experimental approaches of determining the structure of nucleic acids, such as RNA and DNA, can be largely classified into biophysical and biochemical methods. Biophysical methods use the fundamental physical properties of molecules for structure determination, including X-ray crystallography, NMR and cryo-EM. Biochemical methods exploit the chemical properties of nucleic acids using specific reagents and conditions to assay the structure of nucleic acids.[1] Such methods may involve chemical probing with specific reagents, or rely on native or analogue chemistry. Different experimental approaches have unique merits and are suitable for different experimental purposes.
Biophysical methods
[edit]X-ray crystallography
[edit]This section needs expansion with: its meaningful contribution to the study of nucleic acid structures, and some examples. You can help by adding to it. (December 2018) |
X-ray crystallography is not common for nucleic acids alone, since neither DNA nor RNA readily form crystals. This is due to the greater degree of intrinsic disorder and dynamism in nucleic acid structures and the negatively charged (deoxy)ribose-phosphate backbones, which repel each other in close proximity. Therefore, crystallized nucleic acids tend to be complexed with a protein of interest to provide structural order and neutralize the negative charge.[citation needed]
Nuclear magnetic resonance spectroscopy (NMR)
[edit]Nucleic acid NMR is the use of NMR spectroscopy to obtain information about the structure and dynamics of nucleic acid molecules, such as DNA or RNA. As of 2003, nearly half of all known RNA structures had been determined by NMR spectroscopy.[2]
Nucleic acid NMR uses similar techniques as protein NMR, but has several differences. Nucleic acids have a smaller percentage of hydrogen atoms, which are the atoms usually observed in NMR, and because nucleic acid double helices are stiff and roughly linear, they do not fold back on themselves to give "long-range" correlations.[3] The types of NMR usually done with nucleic acids are 1H or proton NMR, 13C NMR, 15N NMR, and 31P NMR. Two-dimensional NMR methods are almost always used, such as correlation spectroscopy (COSY) and total coherence transfer spectroscopy (TOCSY) to detect through-bond nuclear couplings, and nuclear Overhauser effect spectroscopy (NOESY) to detect couplings between nuclei that are close to each other in space.[4]
Parameters taken from the spectrum, mainly NOESY cross-peaks and coupling constants, can be used to determine local structural features such as glycosidic bond angles, dihedral angles (using the Karplus equation), and sugar pucker conformations. For large-scale structure, these local parameters must be supplemented with other structural assumptions or models, because errors add up as the double helix is traversed, and unlike with proteins, the double helix does not have a compact interior and does not fold back upon itself. NMR is also useful for investigating nonstandard geometries such as bent helices, non-Watson–Crick basepairing, and coaxial stacking. It has been especially useful in probing the structure of natural RNA oligonucleotides, which tend to adopt complex conformations such as stem-loops and pseudoknots. NMR is also useful for probing the binding of nucleic acid molecules to other molecules, such as proteins or drugs, by seeing which resonances are shifted upon binding of the other molecule.[4]
Cryogenic electron microscopy (cryo-EM)
[edit]This section needs expansion. You can help by adding to it. (March 2020) |
Cryogenic electron microscopy (cryo-EM) is a technique that uses an electron beam to image samples that have been cryogenically preserved in an aqueous solution. Liquid samples are pipetted on small metallic grids and plunged into a liquid ethane/propane solution which is kept extremely cold by a liquid nitrogen bath. Upon this freezing process, water molecules in the sample do not have enough time to form hexagonal lattices as found in ice, and therefore the sample is preserved in a glassy water-like state (also referred to as a vitrified ice), making these samples easier to image using the electron beam. An advantage of cryo-EM over x-ray crystallography is that the samples are preserved in their aqueous solution state and not perturbed by forming a crystal of the sample. One disadvantage, is that it is difficult to resolve nucleic acid or protein structures that are smaller than ~75 kilodaltons, partly due to the difficulty of having enough contrast to locate particles in this vitrified aqueous solution. Another disadvantage is that to attain atomic-level structure information about a sample requires taking many images (often referred to as electron micrographs) and averaging over those images in a process called single-particle reconstruction. This is a computationally intensive process.
Cryo-EM is a newer, less perturbative version of transmission electron microscopy (TEM). It is less perturbative because the sample is not dried onto a surface, this drying process is often done in negative-stain TEM, and because Cryo-EM does not require contrast agent like heavy metal salts (e.g. uranyl acetate or phoshotungstic acid) which also may affect the structure of the biomolecule. Transmission electron microscopy, as a technique, utilizes the fact that samples interact with a beam of electrons and only parts of the sample that do not interact with the electron beam are allowed to 'transmit' onto the electron detection system. TEM, in general, has been a useful technique in determining nucleic acid structure since the 1960s.[5][6] While double-stranded DNA (dsDNA) structure may not traditionally be considered structure, in the typical sense of alternating segments of single- and double-stranded regions, in reality, dsDNA is not simply a perfectly ordered double helix at every location of its length due to thermal fluctuations in the DNA and alternative structures that can form like g-quadruplexes. CryoEM of nucleic acid has been done on ribosomes,[7] viral RNA,[8] and single-stranded RNA structures within viruses.[9][10] These studies have resolved structural features at different resolutions from the nucleobase level (2-3 angstroms) up to tertiary structure motifs (greater than a nanometer).
Chemical probing
[edit]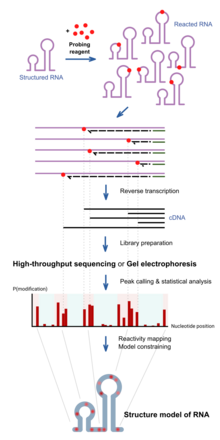
RNA chemical probing uses chemicals that react with RNAs. Importantly, their reactivity depends on local RNA structure e.g. base-pairing or accessibility. Differences in reactivity can therefore serve as a footprint of structure along the sequence. Different reagents react at different positions on the RNA structure, and have different spectra of reactivity.[1] Recent advances allow the simultaneous study of the structure of many RNAs (transcriptome-wide probing)[11] and the direct assay of RNA molecules in their cellular environment (in-cell probing).[12]
Structured RNA is first reacted with the probing reagents for a given incubation time. These reagents would form a covalent adduct on the RNA at the site of reaction. When the RNA is reverse transcribed using a reverse transcriptase into a DNA copy, the DNA generated is truncated at the positions of reaction because the enzyme is blocked by the adducts. The collection of DNA molecules of various truncated lengths therefore informs the frequency of reaction at every base position, which reflects the structure profile along the RNA. This is traditionally assayed by running the DNA on a gel, and the intensity of bands inform the frequency of observing a truncation at each position. Recent approaches use high-throughput sequencing to achieve the same purpose with greater throughput and sensitivity.
The reactivity profile can be used to study the degree of structure at particular positions for specific hypotheses, or used in conjunction with computational algorithms to produce a complete experimentally supported structure model.[13]
Depending on the chemical reagent used, some reagents, e.g. hydroxyl radicals, would cleave the RNA molecule instead. The result in the truncated DNA is the same. Some reagents, e.g. DMS, sometimes do not block the reverse transcriptase, but trigger a mistake at the site in the DNA copy instead. These can be detected when using high-throughput sequencing methods, and is sometimes employed for improved results of probing as mutational profiling (MaP).[14][15]
Positions on the RNA can be protected from the reagents not only by local structure but also by a binding protein over that position. This has led some work to use chemical probing to also assay protein-binding.[16]
Hydroxyl radical probing
[edit]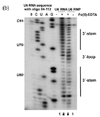
As hydroxyl radicals are short-lived in solution, they need to be generated upon experiment. This can be done using H2O2, ascorbic acid, and Fe(II)-EDTA complex. These reagents form a system that generates hydroxyl radicals through Fenton chemistry. The hydroxyl radicals can then react with the nucleic acid molecules.[17] Hydroxyl radicals attack the ribose/deoxyribose ring and this results in breaking of the sugar-phosphate backbone. Sites under protection from binding proteins or RNA tertiary structure would be cleaved by hydroxyl radical at a lower rate.[17] These positions would therefore show up as absence of bands on the gel, or low signal through sequencing.[17][18]
DMS
[edit]Dimethyl sulfate, known as DMS, is a chemical that can be used to modify nucleic acids in order to determine secondary structure. Reaction with DMS adds a methyl adduct at the site, known as methylation. In particular, DMS methylates N1 of adenine (A) and N3 of cytosine (C),[19] both located at the site of natural hydrogen bonds upon base-pairing. Therefore, modification can only occur at A and C nucleobases that are single-stranded, base paired at the end of a helix, or in a base pair at or next to a GU wobble pair, the latter two being positions where the base-pairing can occasionally open up. Moreover, since modified sites cannot be base-paired, modification sites can be detected by RT-PCR, where the reverse transcriptase falls off at methylated bases and produces different truncated cDNAs. These truncated cDNAs can be identified through gel electrophoresis or high-throughput sequencing.
Improving upon truncation-based methods, DMS mutational profiling with sequencing (DMS-MaPseq) can detect multiple DMS modifications in a single RNA molecule, which enables one to obtain more information per read (for a read of 150 nt, typically two to three mutation sites, rather than zero to one truncation sites), determine structures of low-abundance RNAs, and identify subpopulations of RNAs with alternative secondary structures.[20] DMS-MaPseq uses a thermostable group II intron reverse transcriptase (TGIRT) that creates a mutation (rather than a truncation) in the cDNA when it encounters a base methylated by DMS, but otherwise it reverse transcribes with high fidelity. Sequencing the resulting cDNA identifies which bases were mutated during reverse transcription; these bases cannot have been base-paired in the original RNA.
DMS modification can also be used for DNA, for example in footprinting DNA-protein interactions.[21]
SHAPE
[edit]Selective 2′-hydroxyl acylation analyzed by primer extension, or SHAPE, takes advantage of reagents that preferentially modify the backbone of RNA in structurally flexible regions.

Reagents such as N-methylisatoic anhydride (NMIA) and 1-methyl-7-nitroisatoic anhydride (1M7) [22] react with the 2'-hydroxyl group to form adducts on the 2'-hydroxyl of the RNA backbone. Compared to the chemicals used in other RNA probing techniques, these reagents have the advantage of being largely unbiased to base identity, while remaining very sensitive to conformational dynamics. Nucleotides which are constrained (usually by base-pairing) show less adduct formation than nucleotides which are unpaired. Adduct formation is quantified for each nucleotide in a given RNA by extension of a complementary DNA primer with reverse transcriptase and comparison of the resulting fragments with those from an unmodified control.[23] SHAPE therefore reports on RNA structure at the individual nucleotide level. This data can be used as input to generate highly accurate secondary structure models.[24] SHAPE has been used to analyze diverse RNA structures, including that of an entire HIV-1 genome.[25] The best approach is to use a combination of chemical probing reagents and experimental data.[26] In SHAPE-Seq SHAPE is extended by bar-code based multiplexing combined with RNA-Seq and can be performed in a high-throughput fashion.[27]
Carbodiimides
[edit]

The carbodiimide moiety can also form covalent adducts at exposed nucleobases, which are uracil, and to a smaller extent guanine, upon nucleophilic attack by a deprotonated N. They react primarily with N3 of uracil and N1 of guanine modifying two sites responsible for hydrogen bonding on the bases.[19]
1-cyclohexyl-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate, also known as CMCT or CMC, is the most commonly used carbodiimide for RNA structure probing.[29][30] Similar to DMS, it can be detected by reverse transcription followed by gel electrophoresis or high-throughput sequencing. As it is reactive towards G and U, it can be used to complement the data from DMS probing experiments, which inform A and C.[31]
1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, also known as EDC, is a water-soluble carbodiimide that exhibits similar reactivity as CMC, and is also used for the chemical probing of RNA structure. EDC is able to permeate into cells and is thus used for direct in-cell probing of RNA in their native environments.[32][28]
Kethoxal, glyoxal and derivatives
[edit]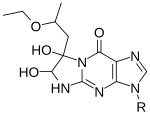

Some 1,2-dicarbonyl compounds are able to react with single-stranded guanine (G) at N1 and N2, forming a five-membered ring adduct at the Watson-Crick face.
1,1-Dihydroxy-3-ethoxy-2-butanone, also known as kethoxal, has a structure related to 1,2-dicarbonyls, and was the first in this category used extensively for the chemical probing of RNA. Kethoxal causes the modification of guanine, specifically altering the N1 and the exocyclic amino group (N2) simultaneously by covalent interaction.[35]
Glyoxal, methylglyoxal, and phenylglyoxal, which all carry the key 1,2-dicarbonyl moiety, all react with free guanines similar to kethoxal, and can be used to probe unpaired guanine bases in structured RNA. Due to their chemical properties, these reagents can permeate readily into cells and can therefore be used to assay RNAs in their native cellular environments.[34]
LASER or NAz Probing
[edit]Light-Activated Structural Examination of RNA (LASER) probing utilizes UV light to activate nicotinoyl azide (NAz), generating highly reactive nitrenium cation in water, which reacts with solvent accessible guanosine and adenosine of RNA at C-8 position through a barrierless Friedel-Crafts reaction. LASER probing targets both single-stranded and double-stranded residues as long as they are solvent accessible. Because hydroxyl radical probing requires synchrotron radiation to measure solvent accessibility of RNA in vivo, it is hard to apply hydroxyl radical probing to footprint RNA in cells for many laboratories. In contrast, LASER probing utilizes a hand-held UV lamp (20 W) for excitation, it is much easier to apply LASER probing for in vivo studying RNA solvent accessibility. This chemical probing method is light-controllable, and probes solvent accessibility of nucleobase, which has been shown to footprint RNA binding proteins inside cells.[36]
In-line probing
[edit]
In-line probing does not involve treatment with any type of chemical or reagent to modify RNA structures. This type of probing assay uses the structure dependent cleavage of RNA; single stranded regions are more flexible and unstable and will degrade over time.[38] The process of in-line probing is often used to determine changes in structure due to ligand binding. Binding of a ligand can result in different cleavage patterns. The process of in-line probing involves incubation of structural or functional RNAs over a long period of time. This period can be several days, but varies in each experiment. The incubated products are then run on a gel to visualize the bands. This experiment is often done using two different conditions: 1) with ligand and 2) in the absence of ligand.[37] Cleavage results in shorter band lengths and is indicative of areas that are not basepaired, as basepaired regions tend to be less sensitive to spontaneous cleavage.[38] In-line probing is a functional assay that can be used to determine structural changes in RNA in response to ligand binding. It can directly show the change in flexibility and binding of regions of RNA in response to a ligand, as well as compare that response to analogous ligands. This assay is commonly used in dynamic studies, specifically when examining riboswitches.[38]
Nucleotide analog interference mapping (NAIM)
[edit]Nucleotide analog interference mapping (NAIM) is the process of using nucleotide analogs, molecules that are similar in some ways to nucleotides but lack function, to determine the importance of a functional group at each location of an RNA molecule.[39][40] The process of NAIM is to insert a single nucleotide analog into a unique site. This can be done by transcribing a short RNA using T7 RNA polymerase, then synthesizing a short oligonucleotide containing the analog in a specific position, then ligating them together on the DNA template using a ligase.[39] The nucleotide analogs are tagged with a phosphorothioate, the active members of the RNA population are then distinguished from the inactive members, the inactive members then have the phosphorothioate tag removed and the analog sites are identified using gel electrophoresis and autoradiography.[39] This indicates a functionally important nucleotide, as cleavage of the phosphorothioate by iodine results in an RNA that is cleaved at the site of the nucleotide analog insert. By running these truncated RNA molecules on a gel, the nucleotide of interest can be identified against a sequencing experiment[40] Site directed incorporation results indicate positions of importance where when running on a gel, functional RNAs that have the analog incorporated at that position will have a band present, but if the analog results in non-functionality, when the functional RNA molecules are run on a gel there will be no band corresponding to that position on the gel.[41] This process can be used to evaluate an entire area, where analogs are placed in site specific locations, differing by a single nucleotide, then when functional RNAs are isolated and run on a gel, all areas where bands are produced indicate non-essential nucleotides, but areas where bands are absent from the functional RNA indicate that inserting a nucleotide analog in that position caused the RNA molecule to become non-functional[39]
References
[edit]- ^ a b Weeks, Kevin (2010). "Advances in RNA structure analysis by chemical probing". Current Opinion in Structural Biology. 20 (3): 295–304. doi:10.1016/j.sbi.2010.04.001. PMC 2916962. PMID 20447823.
- ^ Fürtig B, Richter C, Wöhnert J, Schwalbe H (October 2003). "NMR spectroscopy of RNA". ChemBioChem. 4 (10): 936–62. doi:10.1002/cbic.200300700. PMID 14523911. S2CID 33523981.
- ^ Addess, Kenneth J.; Feigon, Juli (1996). "Introduction to 1H NMR Spectroscopy of DNA". In Hecht, Sidney M. (ed.). Bioorganic Chemistry: Nucleic Acids. New York: Oxford University Press. ISBN 0-19-508467-5.
- ^ a b Wemmer, David (2000). "Chapter 5: Structure and Dynamics by NMR". In Bloomfield, Victor A.; Crothers, Donald M.; Tinoco, Ignacio (eds.). Nucleic acids: Structures, Properties, and Functions. Sausalito, California: University Science Books. ISBN 0-935702-49-0.
- ^ [ GOMATOS PJ, STOECKENIUS W. ELECTRON MICROSCOPE STUDIES ON REOVIRUS RNA. Proceedings of the National Academy of Sciences of the United States of America. 1964 Dec;52:1449-1455. DOI: 10.1073/pnas.52.6.1449]
- ^ [ Michael Beer and Richard Zobel (1961) "Electron stains II: Electron microscopic studies on the visibility of stained DNA molecules" J. Mol. Biol. Volume 3, Issue 6, December 1961, Pages 717–726, IN3–IN5"]
- ^ Frank, J; Penczek, P; Grassucci, R; Srivastava, S (Nov 1991). "Three-dimensional reconstruction of the 70S Escherichia coli ribosome in ice: the distribution of ribosomal RNA". The Journal of Cell Biology. 115 (3): 597–605. doi:10.1083/jcb.115.3.597. PMC 2289182. PMID 1918155.
- ^ Gopal, A; Zhou, ZH; Knobler, CM; Gelbart, WM (2012). "Visualizing large RNA molecules in solution". RNA. 18 (2): 284–299. doi:10.1261/rna.027557.111. PMC 3264915. PMID 22190747.
- ^ Koning, R., Gomez-Blanco, J., Akopjana, I. et al. Asymmetric cryo-EM reconstruction of phage MS2 reveals genome structure in situ. Nat Commun 7, 12524 (2016). https://doi.org/10.1038/ncomms12524
- ^ Dai, X.; Li, Z.; Lai, M.; et al. (2017). "In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus". Nature. 541 (7635): 112–116. Bibcode:2017Natur.541..112D. doi:10.1038/nature20589. PMC 5701785. PMID 27992877.
- ^ Kwok, Chun Kit; Tang, Yin; Assmann, Sarah; Bevilacqua, Philip (April 2015). "The RNA structurome: transcriptome-wide structure probing with next-generation sequencing". Trends in Biochemical Sciences. 40 (4): 221–232. doi:10.1016/j.tibs.2015.02.005. PMID 25797096.
- ^ Kubota, M; Tran, C; Spitale, R (2015). "Progress and challenges for chemical probing of RNA structure inside living cells". Nature Chemical Biology. 11 (12): 933–941. doi:10.1038/nchembio.1958. PMC 5068366. PMID 26575240.
- ^ Mathews, DH; Disney, MD; Childs, JL; Schroeder, SJ; Zuker, M; Turner DH (2004). "Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure". Proceedings of the National Academy of Sciences. 101 (19): 7287–7292. Bibcode:2004PNAS..101.7287M. doi:10.1073/pnas.0401799101. PMC 409911. PMID 15123812.
- ^ Siegfried, N; Busan, S; Weeks, K (2014). "RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP)". Nature Methods. 11 (9): 959–965. doi:10.1038/nmeth.3029. PMC 4259394. PMID 25028896.
- ^ Sexton, A; Wang, P; Rutenberg-Schoenberg, M; Simon, M (2017). "Interpreting Reverse Transcriptase Termination and Mutation Events for Greater Insight into the Chemical Probing of RNA". Biochemistry. 56 (35): 4713–3721. doi:10.1021/acs.biochem.7b00323. PMC 5648349. PMID 28820243.
- ^ Smola, M; Calabrese, J; Weeks, K (2015). "Detection of RNA–Protein Interactions in Living Cells with SHAPE". Biochemistry. 54 (46): 6867–6875. doi:10.1021/acs.biochem.5b00977. PMC 4900165. PMID 26544910.
- ^ a b c d Karaduman R, Fabrizio P, Hartmuth K, Urlaub H, Luhrmann R (2006). "RNA structure and RNA-protein interactions in purified yeast U6 snRNPs". J. Mol. Biol. 356 (5): 1248–1262. doi:10.1016/j.jmb.2005.12.013. hdl:11858/00-001M-0000-0012-E5F8-4. PMID 16410014.
- ^ Tullius, T. D.; Dombroski, B. A. (1986). "Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein". Proceedings of the National Academy of Sciences. 83 (15): 5469–5473. Bibcode:1986PNAS...83.5469T. doi:10.1073/pnas.83.15.5469. PMC 386308. PMID 3090544.
- ^ a b Tijerina P, Mohr S, Russell R (2007). "DMS footprinting of structured RNAs and RNA-protein complexes". Nat Protoc. 2 (10): 2608–23. doi:10.1038/nprot.2007.380. PMC 2701642. PMID 17948004.
- ^ Zubradt, Meghan; Gupta, Paromita; Persad, Sitara; Lambowitz, Alan; Weissman, Jonathan; Rouskin, Silvi (2017). "DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo". Nature Methods. 14 (1): 75–82. doi:10.1038/nmeth.4057. PMC 5508988. PMID 27819661.
- ^ Albert S. Baldwin Jr.; Marjorie Oettinger & Kevin Struhl (1996). "Unit 12.3: Methylation and Uracil Interference Assays for Analysis of Protein-DNA Interactions". Current Protocols in Molecular Biology. Vol. Chapter 12. Wiley. pp. Unit 12.3. doi:10.1002/0471142727.mb1203s36. PMID 18265086. S2CID 45581978.
- ^ Mortimer SA, Weeks KM (2007). "A Fast-Acting Reagent for Accurate Analysis of RNA Secondary and Tertiary Structure by SHAPE Chemistry". J Am Chem Soc. 129 (14): 4144–45. Bibcode:2007JAChS.129.4144M. doi:10.1021/ja0704028. PMID 17367143.
- ^ Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM (2005). "RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE)". J Am Chem Soc. 127 (12): 4223–31. Bibcode:2005JAChS.127.4223M. doi:10.1021/ja043822v. PMID 15783204.
- ^ Deigan KE, Li TW, Mathews DH, Weeks KM (2009). "Accurate SHAPE-directed RNA structure determination". Proc Natl Acad Sci USA. 106 (1): 97–102. Bibcode:2009PNAS..106...97D. doi:10.1073/pnas.0806929106. PMC 2629221. PMID 19109441.
- ^ Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW Jr, Swanstrom R, Burch CL, Weeks KM (2009). "Architecture and secondary structure of an entire HIV-1 RNA genome". Nature. 460 (7256): 711–6. Bibcode:2009Natur.460..711W. doi:10.1038/nature08237. PMC 2724670. PMID 19661910.
- ^ Wipapat Kladwang; Christopher C. VanLang; Pablo Cordero; Rhiju Das (7 Sep 2011). "Understanding the errors of SHAPE-directed RNA structure modeling". Biochemistry. 50 (37): 8049–8056. arXiv:1103.5458. Bibcode:2011arXiv1103.5458K. doi:10.1021/bi200524n. PMC 3172344. PMID 21842868.
- ^ Lucks JB, Mortimer SA, Trapnell C, Luo S, Aviran S, Schroth GP, Pachter L, Doudna JA, Arkin AP (2011). "Multiplexed RNA structure characterization with selective 2'-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq)". Proc Natl Acad Sci USA. 108 (27): 11063–8. Bibcode:2011PNAS..10811063L. doi:10.1073/pnas.1106501108. PMC 3131332. PMID 21642531.
- ^ a b Wang, PY; Sexton, AN; Culligan, WJ; Simon, MD (2019). "Carbodiimide reagents for the chemical probing of RNA structure in cells". RNA. 25 (1): 135–146. doi:10.1261/rna.067561.118. PMC 6298570. PMID 30389828.
- ^ Fritz JJ, Lewin A, Hauswirth W, Agarwal A, Grant M, Shaw L (2002). "Development of hammerhead ribozymes to modulate endogenous gene expression for functional studies". Methods. 28 (2): 276–285. doi:10.1016/S1046-2023(02)00233-5. PMID 12413427.
- ^ Metz, D; Brown, G (1969). "Investigation of nucleic acid secondary structure by means of chemical modification with a carbodiimide reagent. II. Reaction between N-cyclohexyl-N'-β-(4-methylmorpholinium) ethylcarbodiimide and transfer ribonucleic acid". Biochemistry. 8 (6): 2329–2342. doi:10.1021/bi00834a013. PMID 4895018.
- ^ Incarnato, D; Neri, F; Anselmi, F; Oliviero, S (2014). "Genome-wide profiling of mouse RNA secondary structures reveals key features of the mammalian transcriptome". Genome Biology. 15 (491): 491. doi:10.1186/s13059-014-0491-2. PMC 4220049. PMID 25323333.
- ^ Mitchell, D; Renda, A; Douds, C; Babitzke, P; Assmann, S; Bevilacqua, P (2019). "In vivo RNA structural probing of uracil and guanine base-pairing by 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC)". RNA. 25 (1): 147–157. doi:10.1261/rna.067868.118. PMC 6298566. PMID 30341176.
- ^ Noller HF, Chaires JB (1972). "Functional modification of 16S ribosomal RNA by kethoxal". Proc. Natl. Acad. Sci. USA. 69 (11): 3115–3118. Bibcode:1972PNAS...69.3115N. doi:10.1073/pnas.69.11.3115. PMC 389716. PMID 4564202.
- ^ a b Mitchell, D; Ritchey, L; Park, H; Babitzke, P; Assmann, S; Bevilacqua, P (2018). "Glyoxals as in vivo RNA structural probes of guanine base-pairing". RNA. 24 (1): 114–124. doi:10.1261/rna.064014.117. PMC 5733565. PMID 29030489.
- ^ Litt, M; Hancock, V (1967). "Kethoxal—a potentially useful reagent for the determination of nucleotide sequences in single-stranded regions of transfer ribonucleic acid". Biochemistry. 6 (6): 1848–1854. doi:10.1021/bi00858a036. PMID 6035923.
- ^ Feng C, Chan D, Joseph J, Muuronen M, Coldren WH, Dai N, Correa Jr IR, Furche F, Hadad CM, Spitale RC (2018). "Light-activated chemical probing of nucleobase solvent accessibility inside cells". Nat Chem Biol. 14 (3): 276–283. doi:10.1038/nchembio.2548. PMC 6203945. PMID 29334380.
- ^ a b Muhlbacher J, Lafontaine DA (2007). "Ligand recognition determinants of guanine riboswitches". Nucleic Acids Research. 35 (16): 5568–5580. doi:10.1093/nar/gkm572. PMC 2018637. PMID 17704135.
- ^ a b c Regulski, E; Breaker, R (2008). "In-Line Probing Analysis of Riboswitches". In Wilusz, J (ed.). Post-Transcriptional Gene Regulation. Methods in Molecular Biology. Vol. 419. Totowa, NJ: Humana Press. pp. 53–67. doi:10.1007/978-1-59745-033-1_4. ISBN 978-1-58829-783-9. PMID 18369975.
- ^ a b c d Ryder SP, Strobel SA (1999). "Nucleotide Analog Interference Mapping". Methods. 18 (1): 38–50. doi:10.1006/meth.1999.0755. PMID 10208815.
- ^ a b Waldsich C (2008). "Dissecting RNA folding by nucleotide analog interference mapping (NAIM)". Nature Protocols. 3 (5): 811–823. doi:10.1038/nprot.2008.45. PMC 2873565. PMID 18451789.
- ^ Strobel SA, Shetty K (1997). "Defining the chemical groups essential for Tetrahymena group I intron function by nucleotide analog interference mapping". Proc. Natl. Acad. Sci. USA. 94 (7): 2903–2908. Bibcode:1997PNAS...94.2903S. doi:10.1073/pnas.94.7.2903. PMC 20295. PMID 9096319.
