O-GlcNAc

O-GlcNAc (short for O-linked GlcNAc or O-linked β-N-acetylglucosamine) is a reversible enzymatic post-translational modification that is found on serine and threonine residues of nucleocytoplasmic proteins. The modification is characterized by a β-glycosidic bond between the hydroxyl group of serine or threonine side chains and N-acetylglucosamine (GlcNAc). O-GlcNAc differs from other forms of protein glycosylation: (i) O-GlcNAc is not elongated or modified to form more complex glycan structures, (ii) O-GlcNAc is almost exclusively found on nuclear and cytoplasmic proteins rather than membrane proteins and secretory proteins, and (iii) O-GlcNAc is a highly dynamic modification that turns over more rapidly than the proteins which it modifies. O-GlcNAc is conserved across metazoans.[1]

Due to the dynamic nature of O-GlcNAc and its presence on serine and threonine residues, O-GlcNAcylation is similar to protein phosphorylation in some respects. While there are roughly 500 kinases and 150 phosphatases that regulate protein phosphorylation in humans, there are only 2 enzymes that regulate the cycling of O-GlcNAc: O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) catalyze the addition and removal of O-GlcNAc, respectively.[2] OGT utilizes UDP-GlcNAc as the donor sugar for sugar transfer.[3]
First reported in 1984, this post-translational modification has since been identified on over 5,000 proteins.[4][5] Numerous functional roles for O-GlcNAcylation have been reported including crosstalking with serine/threonine phosphorylation, regulating protein-protein interactions, altering protein structure or enzyme activity, changing protein subcellular localization, and modulating protein stability and degradation.[1][6] Numerous components of the cell's transcription machinery have been identified as being modified by O-GlcNAc, and many studies have reported links between O-GlcNAc, transcription, and epigenetics.[7][8] Many other cellular processes are influenced by O-GlcNAc such as apoptosis, the cell cycle, and stress responses.[9] As UDP-GlcNAc is the final product of the hexosamine biosynthetic pathway, which integrates amino acid, carbohydrate, fatty acid, and nucleotide metabolism, it has been suggested that O-GlcNAc acts as a "nutrient sensor" and responds to the cell's metabolic status.[10] Dysregulation of O-GlcNAc has been implicated in many pathologies including Alzheimer's disease, cancer, diabetes, and neurodegenerative disorders.[11][12]
Discovery
[edit]
In 1984, the Hart lab was probing for terminal GlcNAc residues on the surfaces of thymocytes and lymphocytes. Bovine milk β-1,4-galactosyltransferase, which reacts with terminal GlcNAc residues, was used to perform radiolabeling with UDP-[3H]galactose. β-elimination of serine and threonine residues demonstrated that most of the [3H]galactose was attached to proteins O-glycosidically; chromatography revealed that the major β-elimination product was Galβ1-4GlcNAcitol. Insensitivity to peptide N-glycosidase treatment provided additional evidence for O-linked GlcNAc. Permeabilizing cells with detergent prior to radiolabeling greatly increased the amount of [3H]galactose incorporated into Galβ1-4GlcNAcitol, leading the authors to conclude that most of the O-linked GlcNAc monosaccharide residues were intracellular.[13]
Mechanism
[edit]
O-GlcNAc is generally a dynamic modification that can be cycled on and off various proteins. Some residues are thought to be constitutively modified by O-GlcNAc.[14][15] The O-GlcNAc modification is installed by OGT in a sequential bi-bi mechanism where the donor sugar, UDP-GlcNAc, binds to OGT first followed by the substrate protein.[16] The O-GlcNAc modification is removed by OGA in a hydrolysis mechanism involving anchimeric assistance (substrate-assisted catalysis) to yield the unmodified protein and GlcNAc.[17] While crystal structures have been reported for both OGT[16] and OGA,[18][19] the exact mechanisms by which OGT and OGA recognize substrates have not been completely elucidated. Unlike N-linked glycosylation, for which glycosylation occurs in a specific consensus sequence (Asn-X-Ser/Thr, where X is any amino acid except Pro), no definitive consensus sequence has been identified for O-GlcNAc,. Consequently, predicting sites of O-GlcNAc modification is challenging, and identifying modification sites generally requires mass spectrometry methods. For OGT, studies have shown that substrate recognition is regulated by a number of factors including aspartate[20] and asparagine[21] ladder motifs in the lumen of the superhelical TPR domain, active site residues,[22] and adaptor proteins.[23] As crystal structures have shown that OGT requires its substrate to be in an extended conformation, it has been proposed that OGT has a preference for flexible substrates.[22] In in vitro kinetic experiments measuring OGT and OGA activity on a panel of protein substrates, kinetic parameters for OGT were shown to be variable between various proteins while kinetic parameters for OGA were relatively constant between various proteins. This result suggested that OGT is the "senior partner" in regulating O-GlcNAc and OGA primarily recognizes substrates via the presence of O-GlcNAc rather than the identity of the modified protein.[14]

Detection and characterization
[edit]Several methods exist to detect the presence of O-GlcNAc and characterize the specific residues modified.
Lectins
[edit]Wheat germ agglutinin, a plant lectin, is able to recognize terminal GlcNAc residues and is thus often used for detection of O-GlcNAc. This lectin has been applied in lectin affinity chromatography for the enrichment and detection of O-GlcNAc.[24]
Antibodies
[edit]Pan-O-GlcNAc antibodies that recognize the O-GlcNAc modification largely irrespective of the modified protein's identity are commonly used. These include RL2,[25] an IgG antibody raised against O-GlcNAcylated nuclear pore complex proteins, and CTD110.6,[26] an IgM antibody raised against an immunogenic peptide with a single serine O-GlcNAc modification. Other O-GlcNAc-specific antibodies have been reported and demonstrated to have some dependence on the identity of the modified protein.[27]
Metabolic labeling
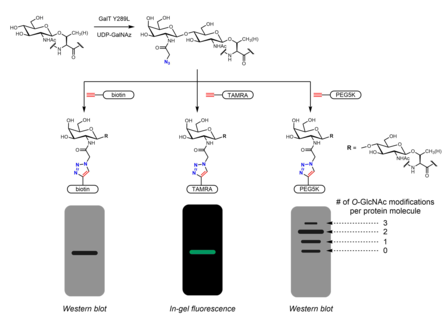
[edit]Many metabolic chemical reporters have been developed to identify O-GlcNAc. Metabolic chemical reporters are generally sugar analogues that bear an additional chemical moiety allowing for additional reactivity. For example, peracetylated GlcNAc (Ac4GlcNAz) is a cell-permeable azido sugar that is de-esterified intracellularly by esterases to GlcNAz and converted to UDP-GlcNAz in the hexosamine salvage pathway. UDP-GlcNAz can be utilized as a sugar donor by OGT to yield the O-GlcNAz modification.[28] The presence of the azido sugar can then be visualized via alkyne-containing bioorthogonal chemical probes in an azide-alkyne cycloaddition reaction. These probes can incorporate easily identifiable tags such as the FLAG peptide, biotin, and dye molecules.[28][29] Mass tags based on polyethylene glycol (PEG) have also been used to measure O-GlcNAc stoichiometry. Conjugation of 5 kDa PEG molecules leads to a mass shift for modified proteins - more heavily O-GlcNAcylated proteins will have multiple PEG molecules and thus migrate more slowly in gel electrophoresis.[30] Other metabolic chemical reporters bearing azides or alkynes (generally at the 2 or 6 positions) have been reported.[31] Instead of GlcNAc analogues, GalNAc analogues may be used as well as UDP-GalNAc is in equilibrium with UDP-GlcNAc in cells due to the action of UDP-galactose-4'-epimerase (GALE). Treatment with Ac4GalNAz was found to result in enhanced labeling of O-GlcNAc relative to Ac4GlcNAz, possibly due to a bottleneck in UDP-GlcNAc pyrophosphorylase processing of GlcNAz-1-P to UDP-GlcNAz.[32] Ac3GlcN-β-Ala-NBD-α-1-P(Ac-SATE)2, a metabolic chemical reporter that is processed intracellularly to a fluorophore-labeled UDP-GlcNAc analogue, has been shown to achieve one-step fluorescent labeling of O-GlcNAc in live cells.[33]
Metabolic labeling may also be used to identify binding partners of O-GlcNAcylated proteins. The N-acetyl group may be elongated to incorporate a diazirine moiety. Treatment of cells with peracetylated, phosphate-protected Ac3GlcNDAz-1-P(Ac-SATE)2 leads to modification of proteins with O-GlcNDAz. UV irradiation then induces photocrosslinking between proteins bearing the O-GlcNDaz modification and interacting proteins.[34]
Some issues have been identified with various metabolic chemical reporters, e.g., their use may inhibit the hexosamine biosynthetic pathway,[31] they may not be recognized by OGA and therefore are not able to capture O-GlcNAc cycling,[35] or they may be incorporated into glycosylation modifications besides O-GlcNAc as seen in secreted proteins.[36] Metabolic chemical reporters with chemical handles at the N-acetyl position may also label acetylated proteins as the acetyl group may be hydrolyzed into acetate analogues that can be utilized for protein acetylation.[37] Additionally, per-O-acetylated monosaccharides have been identified to react cysteines leading to artificial S-glycosylation.[38] This occurs via an elimination-addition mechanism.[39]
Chemoenzymatic labeling
[edit]Chemoenzymatic labeling provides an alternative strategy to incorporate handles for click chemistry. The Click-IT O-GlcNAc Enzymatic Labeling System, developed by the Hsieh-Wilson group and subsequently commercialized by Invitrogen, utilizes a mutant GalT Y289L enzyme that is able to transfer azidogalactose (GalNAz) onto O-GlcNAc.[29][40] The presence of GalNAz (and therefore also O-GlcNAc) can be detected with various alkyne-containing probes with identifiable tags such as biotin,[40] dye molecules,[29] and PEG.[30]

Förster resonance energy transfer biosensor
[edit]An engineered protein biosensor has been developed that can detect changes in O-GlcNAc levels using Förster resonance energy transfer. This sensor consists of four components linked together in the following order: cyan fluorescent protein (CFP), an O-GlcNAc binding domain (based on GafD, a lectin sensitive for terminal β-O-GlcNAc), a CKII peptide that is a known OGT substrate, and yellow fluorescent protein (YFP). Upon O-GlcNAcylation of the CKII peptide, the GafD domain binds the O-GlcNAc moiety, bringing the CFP and YFP domains into close proximity and generating a FRET signal. Generation of this signal is reversible and can be used to monitor O-GlcNAc dynamics in response to various treatments. This sensor may be genetically encoded and used in cells.[41] Addition of a localization sequence allows for targeting of this O-GlcNAc sensor to the nucleus, cytoplasm, or plasma membrane.[42]
Mass spectrometry
[edit]Biochemical approaches such as Western blotting may provide supporting evidence that a protein is modified by O-GlcNAc; mass spectrometry (MS) is able to provide definitive evidence as to the presence of O-GlcNAc. Glycoproteomic studies applying MS have contributed to the identification of proteins modified by O-GlcNAc.
As O-GlcNAc is substoichiometric and ion suppression occurs in the presence of unmodified peptides, an enrichment step is usually performed prior to mass spectrometry analysis. This may be accomplished using lectins, antibodies, or chemical tagging. The O-GlcNAc modification is labile under collision-induced fragmentation methods such as collision-induced dissociation (CID) and higher-energy collisional dissociation (HCD), so these methods in isolation are not readily applicable for O-GlcNAc site mapping. HCD generates fragment ions characteristic of N-acetylhexosamines that can be used to determine O-GlcNAcylation status.[43] In order to facilitate site mapping with HCD, β-elimination followed by Michael addition with dithiothreitol (BEMAD) may be used to convert the labile O-GlcNAc modification into a more stable mass tag. For BEMAD mapping of O-GlcNAc, the sample must be treated with phosphatatase otherwise other serine/threonine post-translational modifications such as phosphorylation may be detected.[44] Electron-transfer dissociation (ETD) is used for site mapping as ETD causes peptide backbone cleavage while leaving post-translational modifications such as O-GlcNAc intact.[45]
Traditional proteomic studies perform tandem MS on the most abundant species in the full-scan mass spectra, prohibiting full characterization of lower-abundance species. One modern strategy for targeted proteomics uses isotopic labels, e.g., dibromide, to tag O-GlcNAcylated proteins. This method allows for algorithmic detection of low-abundance species, which are then sequenced by tandem MS.[46] Directed tandem MS and targeted glycopeptide assignment allow for identification of O-GlcNAcylated peptide sequences. One example probe consists of a biotin affinity tag, an acid-cleavable silane, an isotopic recoding motif, and an alkyne.[47][48][49] Unambiguous site mapping is possible for peptides with only one serine/threonine residue.[50]
The general procedure for this isotope-targeted glycoproteomics (IsoTaG) method is the following:

- Metabolically label O-GlcNAc to install O-GlcNAz onto proteins
- Use click chemistry to link IsoTaG probe to O-GlcNAz
- Use streptavidin beads to enrich for tagged proteins
- Treat beads with trypsin to release non-modified peptides
- Cleave isotopically recoded glycopeptides from beads using mild acid
- Obtain a full-scan mass spectrum from isotopically recoded glycopeptides
- Apply algorithm to detect unique isotope signature from probe
- Perform tandem MS on the isotopically recoded species to obtain glycopeptide amino acid sequences
- Search protein database for identified sequences
Other methodologies have been developed for quantitative profiling of O-GlcNAc using differential isotopic labeling.[51] Example probes generally consist of a biotin affinity tag, a cleavable linker (acid- or photo-cleavable), a heavy or light isotopic tag, and an alkyne.[52][53]
Strategies for manipulating O-GlcNAc
[edit]Various chemical and genetic strategies have been developed to manipulate O-GlcNAc, both on a proteome-wide basis and on specific proteins.
Chemical methods
[edit]Small molecule inhibitors have been reported for both OGT[54][55] and OGA[56][57] that function in cells or in vivo. OGT inhibitors result in a global decrease of O-GlcNAc while OGA inhibitors result in a global increase of O-GlcNAc; these inhibitors are not able to modulate O-GlcNAc on specific proteins.
Inhibition of the hexosamine biosynthetic pathway is also able to decrease O-GlcNAc levels. For instance, glutamine analogues azaserine and 6-diazo-5-oxo-L-norleucine (DON) can inhibit GFAT, though these molecules may also non-specifically affect other pathways.[58]
Protein synthesis
[edit]Expressed protein ligation has been used to prepare O-GlcNAc-modified proteins in a site-specific manner. Methods exist for solid-phase peptide synthesis incorporation of GlcNAc-modified serine, threonine, or cysteine.[59][60]
Genetic methods
[edit]Site-directed mutagenesis
[edit]
Site-directed mutagenesis of O-GlcNAc-modified serine or threonine residues to alanine may be used to evaluate the function of O-GlcNAc at specific residues. As alanine's side chain is a methyl group and is thus not able to act as an O-GlcNAc site, this mutation effectively permanently removes O-GlcNAc at a specific residue. While serine/threonine phosphorylation may be modeled by mutagenesis to aspartate or glutamate, which have negatively charged carboxylate side chains, none of the 20 canonical amino acids sufficiently recapitulate the properties of O-GlcNAc.[61] Mutagenesis to tryptophan has been used to mimic the steric bulk of O-GlcNAc, though tryptophan is much more hydrophobic than O-GlcNAc.[62][63] Mutagenesis may also perturb other post-translational modifications, e.g., if a serine is alternatively phosphorylated or O-GlcNAcylated, alanine mutagenesis permanently eliminates the possibilities of both phosphorylation and O-GlcNAcylation.
S-GlcNAc
[edit]Mass spectrometry identified S-GlcNAc as a post-translational modification found on cysteine residues. In vitro experiments demonstrated that OGT could catalyze the formation of S-GlcNAc and that OGA is incapable of hydrolyzing S-GlcNAc.[64] Though a previous report suggested that OGA is capable of hydrolyzing thioglycosides, this was only demonstrated on the aryl thioglycoside para-nitrophenol-S-GlcNAc; para-nitrothiophenol is a more activated leaving group than a cysteine residue.[65] Recent studies have supported the use of S-GlcNAc as an enzymatically stable structural model of O-GlcNAc that can be incorporated through solid-phase peptide synthesis or site-directed mutagenesis.[66][61][59][67]
Engineered OGT
[edit]Fusion constructs of a nanobody and TPR-truncated OGT allow for proximity-induced protein-specific O-GlcNAcylation in cells. The nanobody may be directed towards protein tags, e.g., GFP, that are fused to the target protein, or the nanobody may be directed towards endogenous proteins. For example, a nanobody recognizing a C-terminal EPEA sequence can direct OGT enzymatic activity to α-synuclein.[68]
Functions of O-GlcNAc
[edit]Apoptosis
[edit]Apoptosis, a form of controlled cell death, has been suggested to be regulated by O-GlcNAc. In various cancers, elevated O-GlcNAc levels have been reported to suppress apoptosis.[69][70] Caspase-3, caspase-8, and caspase-9 have been reported to be modified by O-GlcNAc. Caspase-8 is modified near its cleavage/activation sites; O-GlcNAc modification may block caspase-8 cleavage and activation by steric hindrance. Pharmacological lowering of O-GlcNAc with 5S-GlcNAc accelerated caspase activation while pharmacological raising of O-GlcNAc with thiamet-G inhibited caspase activation.[63]
Epigenetics
[edit]Writers and Erasers
[edit]The proteins that regulate genetics are often categorized as writers, readers, and erasers, i.e., enzymes that install epigenetic modifications, proteins that recognize these modifications, and enzymes that remove these modifications.[71] To date, O-GlcNAc has been identified on writer and eraser enzymes. O-GlcNAc is found in multiple locations on EZH2, the catalytic methyltransferase subunit of PRC2, and is thought to stabilize EZH2 prior to PRC2 complex formation and regulate di- and tri-methyltransferase activity.[72][73] All three members of the ten-eleven translocation (TET) family of dioxygenases (TET1, TET2, and TET3) are known to be modified by O-GlcNAc.[74] O-GlcNAc has been suggested to cause nuclear export of TET3, reducing its enzymatic activity by depleting it from the nucleus.[75] O-GlcNAcylation of HDAC1 is associated with elevated activating phosphorylation of HDAC1.[76]
Histone O-GlcNAcylation
[edit]Histone proteins, the primary protein component of chromatin, are known to be modified by O-GlcNAc.[8] O-GlcNAc has been identified on all core histones (H2A,[8] H2B,[8] H3,[77] and H4[8]). The presence of O-GlcNAc on histones has been suggested to affect gene transcription as well as other histone marks such as acetylation[8] and monoubiquitination.[78] TET2 has been reported to interact with the TPR domain of OGT and facilitate recruitment of OGT to histones.[79] This interaction is associated with H2B S112 O-GlcNAc, which in turn is associated with H2B K120 monoubiquitination.[78] Phosphorylation of OGT T444 via AMPK has been found to inhibit OGT-chromatin association and downregulate H2B S112 O-GlcNAc.[80]
Nutrient sensing
[edit]The hexosamine biosynthetic pathway's product, UDP-GlcNAc, is utilized by OGT to catalyze the addition of O-GlcNAc. This pathway integrates information about the concentrations of various metabolites including amino acids, carbohydrates, fatty acids, and nucleotides. Consequently, UDP-GlcNAc levels are sensitive to cellular metabolite levels. OGT activity is in part regulated by UDP-GlcNAc concentration, making a link between cellular nutrient status and O-GlcNAc.[81]
Glucose deprivation causes a decline in UDP-GlcNAc levels and an initial decline in O-GlcNAc, but counterintuitively, O-GlcNAc is later significantly upregulated. This later increase has been shown to be dependent on AMPK and p38 MAPK activation, and this effect is partially due to increases in OGT mRNA and protein levels.[82] It has also been suggested that this effect is dependent on calcium and CaMKII.[83] Activated p38 is able to recruit OGT to specific protein targets, including neurofilament H; O-GlcNAc modification of neurofilament H enhances its solubility.[82] During glucose deprivation, glycogen synthase is modified by O-GlcNAc which inhibits its activity.[84]
Oxidative stress
[edit]NRF2, a transcription factor associated with the cellular response to oxidative stress, has been found to be indirectly regulated by O-GlcNAc. KEAP1, an adaptor protein for the cullin 3-dependent E3 ubiquitin ligase complex, mediates the degradation of NRF2; oxidative stress leads to conformational changes in KEAP1 that repress degradation of NRF2. O-GlcNAc modification of KEAP1 at S104 is required for efficient ubiquitination and subsequent degradation of NRF2, linking O-GlcNAc to oxidative stress. Glucose deprivation leads to a reduction in O-GlcNAc and reduces NRF2 degradation. Cells expressing a KEAP1 S104A mutant are resistant to erastin-induced ferroptosis, consistent with higher NRF2 levels upon removal of S104 O-GlcNAc.[85]
Elevated O-GlcNAc levels have been associated with diminished synthesis of hepatic glutathione, an important cellular antioxidant. Acetaminophen overdose leads to accumulation of the strongly oxidizing metabolite NAPQI in the liver, which is detoxified by glutathione. In mice, OGT knockout has a protective effect against acetaminophen-induced liver injury, while OGA inhibition with thiamet-G exacerbates acetaminophen-induced liver injury.[86]
Protein aggregation
[edit]O-GlcNAc has been found to slow protein aggregation, though the generality of this phenomenon is unknown.
Solid-phase peptide synthesis was used to prepare full-length α-synuclein with an O-GlcNAc modification at T72. Thioflavin T aggregation assays and transmission electron microscopy demonstrated that this modified α-synuclein does not readily form aggregates.[60]
Treatment of JNPL3 tau transgenic mice with an OGA inhibitor was shown to increase microtubule-associated protein tau O-GlcNAcylation. Immunohistochemistry analysis of the brainstem revealed decreased formation of neurofibrillary tangles. Recombinant O-GlcNAcylated tau was shown to aggregate slower than unmodified tau in an in vitro thioflavin S aggregation assay. Similar results were obtained for a recombinantly prepared O-GlcNAcylated TAB1 construct versus its unmodified form.[87]
Protein phosphorylation
[edit]Crosstalk
[edit]Many known phosphorylation sites and O-GlcNAcylation sites are nearby each other or overlapping.[50] As protein O-GlcNAcylation and phosphorylation both occur on serine and threonine residues, these post-translational modifications can regulate each other. For example, in CKIIα, S347 O-GlcNAc has been shown to antagonize T344 phosphorylation.[59] Reciprocal inhibition, i.e., phosphorylation inhibition of O-GlcNAcylation and O-GlcNAcylation of phosphorylation, has been observed on other proteins including murine estrogen receptor β,[88] RNA Pol II,[89] tau,[90] p53,[91] CaMKIV,[92] p65,[93] β-catenin,[94] and α-synuclein.[60] Positive cooperativity has also been observed between these two post-translational modifications, i.e., phosphorylation induces O-GlcNAcylation or O-GlcNAcylation induces phosphorylation. This has been demonstrated on MeCP2[30] and HDAC1.[76] In other proteins, e.g., cofilin, phosphorylation and O-GlcNAcylation appear to occur independently of each other.[95]
In some cases, therapeutic strategies are under investigation to modulate O-GlcNAcylation to have a downstream effect on phosphorylation. For instance, elevating tau O-GlcNAcylation may offer therapeutic benefit by inhibiting pathological tau hyperphosphorylation.[96]
Besides phosphorylation, O-GlcNAc has been found to influence other post-translational modifications such as lysine acetylation[93] and monoubiquitination.[78]
Kinases
[edit]Protein kinases are the enzymes responsible for phosphorylation of serine and threonine residues. O-GlcNAc has been identified on over 100 (~20% of the human kinome) kinases, and this modification is often associated with alterations in kinase activity or kinase substrate scope.[97]
The first report of a kinase being directly regulated by O-GlcNAc was published in 2009. CaMKIV is glycosylated at multiple sites, though S189 was found to be the major site. An S189A mutant was more readily activated by CaMKIV T200 phosphorylation, suggesting that O-GlcNAc at S189 inhibits CaMKIV activity. Homology modeling showed that S189 O-GlcNAc may interfere with ATP binding.[92]
AMPK and OGT are known to modify each other, i.e., AMPK phosphorylates OGT and OGT O-GlcNAcylates AMPK. AMPK activation by AICA ribonucleotide is associated with nuclear localization of OGT in differentiated C2C12 mouse skeletal muscle myotubes, resulting in increased nuclear O-GlcNAc. This effect was not observed in proliferating cells and undifferentiated myoblastic cells.[98] AMPK phosphorylation of OGT T444 has been found to block OGT association with chromatin and decrease H2B S112 O-GlcNAc.[80] Overexpression of GFAT, the enzyme that controls glucose flux into the hexosamine biosynthetic pathway, in mouse adipose tissue has been found to lead to AMPK activation and downstream ACC inhibition and elevated fatty acid oxidation. Glucosamine treatment in cultured 3T3L1 adipocytes showed a similar effect.[99] The exact relationship between O-GlcNAc and AMPK has not been completely elucidated as various studies have reported that OGA inhibition inhibits AMPK activation,[98] OGT inhibition also inhibits AMPK activation,[80] upregulating O-GlcNAc by glucosamine treatment activates AMPK,[99] and OGT knockdown activates AMPK;[100] these results suggest that additional indirect communication between AMPK pathways and O-GlcNAc or cell type-specific effects.
CKIIα substrate recognition has been shown to be altered upon S347 O-GlcNAcylation.[59]
Phosphatases
[edit]Protein phosphatase 1 subunits PP1β and PP1γ have been shown to form functional complexes with OGT. A synthetic phosphopeptide was able to be dephosphorylated and O-GlcNAcylated by an OGT immunoprecipitate. This complex has been referred to as a "yin-yang complex" as it replaces a phosphate modification with an O-GlcNAc modification.[101]
MYPT1 is another protein phosphatase subunit that forms complexes with OGT and is itself O-GlcNAcylated. MYPT1 appears to have a role in directing OGT towards specific substrates.[102]
Protein-protein interactions
[edit]O-GlcNAcylation of a protein can alter its interactome. As O-GlcNAc is highly hydrophilic, its presence may disrupt hydrophobic protein-protein interactions. For example, O-GlcNAc disrupts Sp1 interaction with TAFII110,[103] and O-GlcNAc disrupts CREB interaction with TAFII130 and CRTC.[104][105]
Some studies have also identified instances where protein-protein interactions are induced by O-GlcNAc. Metabolic labeling with the diazirine-containing O-GlcNDAz has been applied to identify protein-protein interactions induced by O-GlcNAc.[34] Using a bait glycopeptide based roughly on a consensus sequence for O-GlcNAc, α-enolase, EBP1, and 14-3-3 were identified as potential O-GlcNAc readers. X-ray crystallography showed that 14-3-3 recognized O-GlcNAc through an amphipathic groove that also binds phosphorylated ligands.[106] Hsp70 has also been proposed to act as a lectin to recognize O-GlcNAc.[107] It has been suggested that O-GlcNAc plays a role in the interaction of α-catenin and β-catenin.[94]
Protein stability and degradation
[edit]Co-translational O-GlcNAc has been identified on Sp1 and Nup62. This modification suppresses co-translational ubiquitination and thus protects nascent polypeptides from proteasomal degradation. Similar protective effects of O-GlcNAc on full-length Sp1 have been observed. It is unknown if this pattern is universal or only applicable to specific proteins.[15]
Protein phosphorylation is often used as a mark for subsequent degradation. Tumor suppressor protein p53 is targeted for proteasomal degradation via COP9 signalosome-mediated phosphorylation of T155. O-GlcNAcylation of p53 S149 has been associated with decreased T155 phosphorylation and protection of p53 from degradation.[91] β-catenin O-GlcNAcylation competes with T41 phosphorylation, which signals β-catenin for degradation, stabilizing the protein.[94]
O-GlcNAcylation of the Rpt2 ATPase subunit of the 26S proteasome has been shown to inhibit proteasome activity. Testing various peptide sequences revealed that this modification slows proteasomal degradation of hydrophobic peptides, degradation of hydrophilic peptides does not appear to be affected.[108] This modification has been shown to suppress other pathways that activate the proteasome such as Rpt6 phosphorylation by cAMP-dependent protein kinase.[109]
OGA-S localizes to lipid droplets and has been proposed to locally activate the proteasome to promote remodeling of lipid droplet surface proteins.[110]
Stress response
[edit]Various cellular stress stimuli have been associated with changes in O-GlcNAc. Treatment with hydrogen peroxide, cobalt(II) chloride, UVB light, ethanol, sodium chloride, heat shock, and sodium arsenite, all result in elevated O-GlcNAc. Knockout of OGT sensitizes cells to thermal stress. Elevated O-GlcNAc has been associated with expression of Hsp40 and Hsp70.[111]
Therapeutic relevance
[edit]Alzheimer's disease
[edit]Numerous studies have identified aberrant phosphorylation of tau as a hallmark of Alzheimer's disease.[112] O-GlcNAcylation of bovine tau was first characterized in 1996.[113] A subsequent report in 2004 demonstrated that human brain tau is also modified by O-GlcNAc. O-GlcNAcylation of tau was demonstrated to regulate tau phosphorylation with hyperphosphorylation of tau observed in the brain of mice lacking OGT,[114] which has been associated with the formation of neurofibrillary tangles. Analysis of brain samples showed that protein O-GlcNAcylation is compromised in Alzheimer's disease and paired helical fragment-tau was not recognized by traditional O-GlcNAc detection methods, suggesting that pathological tau has impaired O-GlcNAcylation relative to tau isolated from control brain samples. Elevating tau O-GlcNAcylation was proposed as a therapeutic strategy for reducing tau phosphorylation.[90]
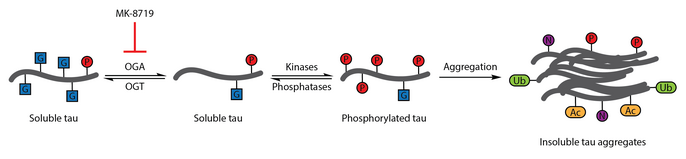
To test this therapeutic hypothesis, a selective and blood-brain barrier-permeable OGA inhibitor, thiamet-G, was developed. Thiamet-G treatment was able to increase tau O-GlcNAcylation and suppress tau phosphorylation in cell culture and in vivo in healthy Sprague-Dawley rats.[57] A subsequent study showed that thiamet-G treatment also increased tau O-GlcNAcylation in a JNPL3 tau transgenic mouse model. In this model, tau phosphorylation was not significantly affected by thiamet-G treatment, though decreased numbers of neurofibrillary tangles and slower motor neuron loss were observed. Additionally, O-GlcNAcylation of tau was noted to slow tau aggregation in vitro.[87]
OGA inhibition with MK-8719 is being investigated in clinical trials as a potential treatment strategy for Alzheimer's disease and other tauopathies including progressive supranuclear palsy.[96][115][116]
Cancer
[edit]Dysregulation of O-GlcNAc is associated with cancer cell proliferation and tumor growth.
O-GlcNAcylation of the glycolytic enzyme PFK1 at S529 has been found to inhibit PFK1 enzymatic activity, reducing glycolytic flux and redirecting glucose towards the pentose phosphate pathway. Structural modeling and biochemical experiments suggested that O-GlcNAc at S529 would inhibit PFK1 allosteric activation by fructose 2,6-bisphosphate and oligomerization into active forms. In a mouse model, mice injected with cells expressing PFK1 S529A mutant showed lower tumor growth than mice injected with cells expressing PFK1 wild-type. Additionally, OGT overexpression enhanced tumor growth in the latter system but had no significant effect on the system with mutant PFK1. Hypoxia induces PFK1 S529 O-GlcNAc and increases flux through the pentose phosphate pathway to generate more NADPH, which maintains glutathione levels and detoxifies reactive oxygen species, imparting a growth advantage to cancer cells. PFK1 was found to be glycosylated in human breast and lung tumor tissues.[117] OGT has also been reported to positively regulate HIF-1α. HIF-1α is normally degraded under normoxic conditions by prolyl hydroxylases that utilize α-ketoglutarate as a co-substrate. OGT suppresses α-ketoglutarate levels, protecting HIF-1α from proteasomal degradation by pVHL and promoting aerobic glycolysis. In contrast with the previous study on PFK1, this study found that elevating OGT or O-GlcNAc upregulated PFK1, though the two studies are consistent in finding that O-GlcNAc levels are positively associated with flux through the pentose phosphate pathway. This study also found that decreasing O-GlcNAc selectively killed cancer cells via ER stress-induced apoptosis.[69]
Human pancreatic ductal adenocarcinoma (PDAC) cell lines have higher O-GlcNAc levels than human pancreatic duct epithelial (HPDE) cells. PDAC cells have some dependency upon O-GlcNAc for survival as OGT knockdown selectively inhibited PDAC cell proliferation (OGT knockdown did not significantly affect HPDE cell proliferation), and inhibition of OGT with 5S-GlcNAc showed the same result. Hyper-O-GlcNAcylation in PDAC cells appeared to be anti-apoptotic, inhibiting cleavage and activation of caspase-3 and caspase-9. Numerous sites on the p65 subunit of NF-κB were found to be modified by O-GlcNAc in a dynamic manner; O-GlcNAc at p65 T305 and S319 in turn positively regulate other modifications associated with NF-κB activation such as p300-mediated K310 acetylation and IKK-mediated S536 phosphorylation. These results suggested that NF-κB is constitutively activated by O-GlcNAc in pancreatic cancer.[70][93]
OGT stabilization of EZH2 in various breast cancer cell lines has been found to inhibit expression of tumor suppressor genes.[72] In hepatocellular carcinoma models, O-GlcNAc is associated with activating phosphorylation of HDAC1, which in turn regulates expression of the cell cycle regulator p21Waf1/Cip1 and cell motility regulator E-cadherin.[76]
OGT has been found to stabilize SREBP-1 and activate lipogenesis in breast cancer cell lines. This stabilization was dependent on the proteasome and AMPK. OGT knockdown resulted in decreased nuclear SREBP-1, but proteasomal inhibition with MG132 blocked this effect. OGT knockdown also increased the interaction between SREBP-1 and the E3 ubiquitin ligase FBW7. AMPK is activated by T172 phosphorylation upon OGT knockdown, and AMPK phosphorylates SREBP-1 S372 to inhibit its cleavage and maturation. OGT knockdown had a diminished effect on SREBP-1 levels in AMPK-null cell lines. In a mouse model, OGT knockdown inhibited tumor growth but SREBP-1 overexpression partly rescued this effect.[100] These results contrast from those of a previous study which found that OGT knockdown/inhibition inhibited AMPK T172 phosphorylation and increased lipogenesis.[80]
In breast and prostate cancer cell lines, high levels of OGT and O-GlcNAc have been associated both in vitro and in vivo with processes associated with disease progression, e.g., angiogenesis, invasion, and metastasis. OGT knockdown or inhibition was found to downregulate the transcription factor FoxM1 and upregulate the cell-cycle inhibitor p27Kip1 (which is regulated by FoxM1-dependent expression of the E3 ubiquitin ligase component Skp2), causing G1 cell cycle arrest. This appeared to be dependent on proteasomal degradation of FoxM1, as expression of a FoxM1 mutant lacking a degron rescued the effects of OGT knockdown. FoxM1 was found not to be directly modified by O-GlcNAc, suggesting that hyper-O-GlcNAcylation of FoxM1 regulators impairs FoxM1 degradation. Targeting OGT also lowered levels of FoxM1-regulated proteins associated with cancer invasion and metastasis (MMP-2 & MMP-9), and angiogenesis (VEGF).[118][119] O-GlcNAc modification of cofilin S108 has also been reported to be important for breast cancer cell invasion by regulating cofilin subcellular localization in invadopodia.[95]
Diabetes
[edit]Elevated O-GlcNAc has been associated with diabetes.
Pancreatic β cells synthesize and secrete insulin to regulate blood glucose levels. One study found that inhibition of OGA with streptozotocin followed by glucosamine treatment resulted in O-GlcNAc accumulation and apoptosis in β cells;[120] a subsequent study showed that a galactose-based analogue of streptozotocin was unable to inhibit OGA but still resulted in apoptosis, suggesting that the apoptotic effects of streptozotocin are not directly due to OGA inhibition.[121]
O-GlcNAc has been suggested to attenuate insulin signaling. In 3T3-L1 adipocytes, OGA inhibition with PUGNAc inhibited insulin-mediated glucose uptake. PUGNAc treatment also inhibited insulin-stimulated Akt T308 phosphorylation and downstream GSK3β S9 phosphorylation.[122] In a later study, insulin stimulation of COS-7 cells caused OGT to localize to the plasma membrane. Inhibition of PI3K with wortmannin reversed this effect, suggesting dependence on phosphatidylinositol(3,4,5)-triphosphate. Increasing O-GlcNAc levels by subjecting cells to high glucose conditions or PUGNAc treatment inhibited insulin-stimulated phosphorylation of Akt T308 and Akt activity. IRS1 phosphorylation at S307 and S632/S635, which is associated with attenuated insulin signaling, was enhanced. Subsequent experiments in mice with adenoviral delivery of OGT showed that OGT overexpression negatively regulated insulin signaling in vivo. Many components of the insulin signaling pathway, including β-catenin,[122] IR-β, IRS1, Akt, PDK1, and the p110α subunit of PI3K were found to be directly modified by O-GlcNAc.[123] Insulin signaling has also been reported to lead to OGT tyrosine phosphorylation and OGT activation, resulting in increased O-GlcNAc levels.[124]
As PUGNAc also inhibits lysosomal β-hexosaminidases, the OGA-selective inhibitor NButGT was developed to further probe the relationship between O-GlcNAc and insulin signaling in 3T3-L1 adipocytes. This study also found that PUGNAc resulted in impaired insulin signaling, but NButGT did not, as measured by changes in phosphorylation of Akt T308, suggesting that the effects observed with PUGNAc may be due to off-target effects besides OGA inhibition.[125]
Parkinson's disease
[edit]Parkinson's disease is associated with aggregation of α-synuclein.[126] As O-GlcNAc modification of α-synuclein has been found to inhibit its aggregation, elevating α-synuclein O-GlcNAc is being explored as a therapeutic strategy to treat Parkinson's disease.[60][127]
Infectious disease
[edit]Bacterial
[edit]Treatment of macrophages with lipopolysaccharide (LPS), a major component of the Gram-negative bacteria outer membrane, results in elevated O-GlcNAc in cellular and mouse models. During infection, cytosolic OGT was de-S-nitrosylated and activated. Suppressing O-GlcNAc with DON inhibited the O-GlcNAcylation and nuclear translocation of NF-κB, as well as downstream induction of inducible nitric oxide synthase and IL-1β production. DON treatment also improved cell survival during LPS treatment.[128]
Viral
[edit]O-GlcNAc has been implicated in influenza A virus (IAV)-induced cytokine storm. Specifically, O-GlcNAcylation of S430 on interferon regulatory factor-5 (IRF5) has been shown to promote its interaction with TNF receptor-associated factor 6 (TRAF6) in cellular and mouse models. TRAF6 mediates K63-linked ubiquitination of IRF5 which is necessary for IRF5 activity and subsequent cytokine production. Analysis of clinical samples showed that blood glucose levels were elevated in IAV-infected patients compared to healthy individuals. In IAV-infected patients, blood glucose levels positively correlated with IL-6 and IL-8 levels. O-GlcNAcylation of IRF5 was also relatively higher in peripheral blood mononuclear cells of IAV-infected patients.[129]
Other applications
[edit]Peptide therapeutics such as are attractive for their high specificity and potency, but they often have poor pharmacokinetic profiles due to their degradation by serum proteases.[130] Though O-GlcNAc is generally associated with intracellular proteins, it has been found that engineered peptide therapeutics modified by O-GlcNAc have enhanced serum stability in a mouse model and have similar structure and activity compared to the respective unmodified peptides. This method has been applied to engineer GLP-1 and PTH peptides.[131]
See also
[edit]References
[edit]- ^ a b Zeidan, Quira; Hart, Gerald W. (2010-01-01). "The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways". Journal of Cell Science. 123 (1): 13–22. doi:10.1242/jcs.053678. ISSN 0021-9533. PMC 2794709. PMID 20016062.
- ^ Dias, Wagner B.; Cheung, Win D.; Hart, Gerald W. (2012-06-01). "O-GlcNAcylation of Kinases". Biochemical and Biophysical Research Communications. 422 (2): 224–228. doi:10.1016/j.bbrc.2012.04.124. ISSN 0006-291X. PMC 3387735. PMID 22564745.
- ^ Haltiwanger, RS; Holt, GD; Hart, GW (1990-02-15). "Enzymatic Addition of O-GlcNAc to Nuclear and Cytoplasmic Proteins. Identification of a Uridine diphospho-N-acetylglucosamine:peptide beta-N-acetylglucosaminyltransferase". Journal of Biological Chemistry. 265 (5): 2563–8. doi:10.1016/S0021-9258(19)39838-2. PMID 2137449.
- ^ Wulff-Fuentes E, Berendt RR, Massman L, Danner L, Malard F, Vora J, Kahsay R, Olivier-Van Stichelen S (January 2021). "The human O-GlcNAcome database and meta-analysis". Scientific Data. 8 (1): 25. Bibcode:2021NatSD...8...25W. doi:10.1038/s41597-021-00810-4. PMC 7820439. PMID 33479245.
- ^ Ma, Junfeng; Hart, Gerald W (2014-03-05). "O-GlcNAc profiling: from proteins to proteomes". Clinical Proteomics. 11 (1): 8. doi:10.1186/1559-0275-11-8. ISSN 1542-6416. PMC 4015695. PMID 24593906.
- ^ King, Dustin T.; Serrano-Negrón, Jesús E.; Zhu, Yanping; Moore, Christopher L.; Shoulders, Matthew D.; Foster, Leonard J.; Vocadlo, David J. (2022-03-09). "Thermal Proteome Profiling Reveals the O-GlcNAc-Dependent Meltome". Journal of the American Chemical Society. 144 (9): 3833–3842. doi:10.1021/jacs.1c10621. ISSN 1520-5126. PMC 8969899. PMID 35230102.
- ^ Kelly, WG; Dahmus, ME; Hart, GW (1993-05-15). "RNA Polymerase II Is a Glycoprotein. Modification of the COOH-terminal Domain by O-GlcNAc". Journal of Biological Chemistry. 268 (14): 10416–24. doi:10.1016/S0021-9258(18)82216-5. PMID 8486697.
- ^ a b c d e f Sakabe, K; Wang, Z; Hart, GW (2010-11-16). "Beta-N-acetylglucosamine (O-GlcNAc) Is Part of the Histone Code". Proceedings of the National Academy of Sciences of the United States of America. 107 (46): 19915–20. Bibcode:2010PNAS..10719915S. doi:10.1073/pnas.1009023107. PMC 2993388. PMID 21045127.
- ^ Levine, Z; Walker, S (2016-06-02). "The Biochemistry of O-GlcNAc Transferase: Which Functions Make It Essential in Mammalian Cells?". Annual Review of Biochemistry. 85: 631–57. doi:10.1146/annurev-biochem-060713-035344. PMID 27294441.
- ^ Ong, Qunxiang; Han, Weiping; Yang, Xiaoyong (2018-10-16). "O-GlcNAc as an Integrator of Signaling Pathways". Frontiers in Endocrinology. 9: 599. doi:10.3389/fendo.2018.00599. ISSN 1664-2392. PMC 6234912. PMID 30464755.
- ^ Cheng, Steven S.; Mody, Alison C.; Woo, Christina M. (2024-11-07). "Opportunities for Therapeutic Modulation of O-GlcNAc". Chemical Reviews. doi:10.1021/acs.chemrev.4c00417. ISSN 0009-2665.
- ^ Hart, Gerald W.; Slawson, Chad; Ramirez-Correa, Genaro; Lagerlof, Olof (2011-06-07). "Cross Talk Between O-GlcNAcylation and Phosphorylation: Roles in Signaling, Transcription, and Chronic Disease". Annual Review of Biochemistry. 80: 825–858. doi:10.1146/annurev-biochem-060608-102511. ISSN 0066-4154. PMC 3294376. PMID 21391816.
- ^ Torres, CR; Hart, GW (1984-03-10). "Topography and Polypeptide Distribution of Terminal N-acetylglucosamine Residues on the Surfaces of Intact Lymphocytes. Evidence for O-linked GlcNAc". Journal of Biological Chemistry. 259 (5): 3308–17. doi:10.1016/S0021-9258(17)43295-9. PMID 6421821.
- ^ a b Shen, David L.; Gloster, Tracey M.; Yuzwa, Scott A.; Vocadlo, David J. (2012-05-04). "Insights into O-Linked N-Acetylglucosamine (O-GlcNAc) Processing and Dynamics through Kinetic Analysis of O-GlcNAc Transferase and O-GlcNAcase Activity on Protein Substrates". Journal of Biological Chemistry. 287 (19): 15395–15408. doi:10.1074/jbc.M111.310664. ISSN 0021-9258. PMC 3346082. PMID 22311971.
- ^ a b Zhu, Y; Liu, TW; Cecioni, S; Eskandari, R; Zandberg, WF; Vocadlo, DJ (May 2015). "O-GlcNAc Occurs Cotranslationally to Stabilize Nascent Polypeptide Chains". Nature Chemical Biology. 11 (5): 319–25. doi:10.1038/nchembio.1774. PMID 25774941.
- ^ a b c Lazarus, MB; Nam, Y; Jiang, J; Sliz, P; Walker, S (2011-01-27). "Structure of Human O-GlcNAc Transferase and Its Complex With a Peptide Substrate". Nature. 469 (7331): 564–7. Bibcode:2011Natur.469..564L. doi:10.1038/nature09638. PMC 3064491. PMID 21240259.
- ^ Macauley, MS; Whitworth, GE; Debowski, AW; Chin, D; Vocadlo, DJ (2005-07-08). "O-GlcNAcase Uses Substrate-Assisted Catalysis: Kinetic Analysis and Development of Highly Selective Mechanism-Inspired Inhibitors". Journal of Biological Chemistry. 280 (27): 25313–22. doi:10.1074/jbc.M413819200. PMID 15795231.
- ^ Roth, Christian; Chan, Sherry; Offen, Wendy A; Hemsworth, Glyn R; Willems, Lianne I; King, Dustin T; Varghese, Vimal; Britton, Robert; Vocadlo, David J; Davies, Gideon J (June 2017). "Structural and functional insight into human O-GlcNAcase". Nature Chemical Biology. 13 (6): 610–612. doi:10.1038/nchembio.2358. ISSN 1552-4450. PMC 5438047. PMID 28346405.
- ^ Elsen, NL; Patel, SB; Ford, RE; Hall, DL; Hess, F; Kandula, H; Kornienko, M; Reid, J; Selnick, H (June 2017). "Insights Into Activity and Inhibition From the Crystal Structure of Human O-GlcNAcase". Nature Chemical Biology. 13 (6): 613–615. doi:10.1038/nchembio.2357. PMID 28346407.
- ^ Joiner, CM; Levine, ZG; Aonbangkhen, C; Woo, CM; Walker, S (2019-08-21). "Aspartate Residues Far From the Active Site Drive O-GlcNAc Transferase Substrate Selection". Journal of the American Chemical Society. 141 (33): 12974–12978. doi:10.1021/jacs.9b06061. PMC 6849375. PMID 31373491.
- ^ Levine, ZG; Fan, C; Melicher, MS; Orman, M; Benjamin, T; Walker, S (2018-03-14). "O-GlcNAc Transferase Recognizes Protein Substrates Using an Asparagine Ladder in the Tetratricopeptide Repeat (TPR) Superhelix". Journal of the American Chemical Society. 140 (10): 3510–3513. doi:10.1021/jacs.7b13546. PMC 5937710. PMID 29485866.
- ^ a b S, Pathak; J, Alonso; M, Schimpl; K, Rafie; De, Blair; Vs, Borodkin; O, Albarbarawi; Dmf, van Aalten (Sep 2015). "The Active Site of O-GlcNAc Transferase Imposes Constraints on Substrate Sequence". Nature Structural & Molecular Biology. 22 (9): 744–750. doi:10.1038/nsmb.3063. PMC 4979681. PMID 26237509.
- ^ Cheung, WD; Sakabe, K; Housley, MP; Dias, WB; Hart, GW (2008-12-05). "O-linked beta-N-acetylglucosaminyltransferase Substrate Specificity Is Regulated by Myosin Phosphatase Targeting and Other Interacting Proteins". Journal of Biological Chemistry. 283 (49): 33935–41. doi:10.1074/jbc.M806199200. PMC 2590692. PMID 18840611.
- ^ Zachara, Natasha E.; Vosseller, Keith; Hart, Gerald W. (November 2011). "Detection and Analysis of Proteins Modified by O-Linked N-Acetylglucosamine". Current Protocols in Protein Science. CHAPTER: 12.8.1–12.8.33. doi:10.1002/0471140864.ps1208s66. ISSN 1934-3655. PMC 3349994. PMID 22045558.
- ^ Snow, C. M.; Senior, A.; Gerace, L. (1987-05-01). "Monoclonal antibodies identify a group of nuclear pore complex glycoproteins". The Journal of Cell Biology. 104 (5): 1143–1156. doi:10.1083/jcb.104.5.1143. ISSN 0021-9525. PMC 2114474. PMID 2437126.
- ^ Comer, FI; Vosseller, K; Wells, L; Accavitti, MA; Hart, GW (2001-06-15). "Characterization of a Mouse Monoclonal Antibody Specific for O-linked N-acetylglucosamine". Analytical Biochemistry. 293 (2): 169–77. doi:10.1006/abio.2001.5132. PMID 11399029.
- ^ Teo, CF; Ingale, S; Wolfert, MA; Elsayed, GA; Nöt, LG; Chatham, JC; Wells, L; Boons, GJ (May 2010). "Glycopeptide-specific Monoclonal Antibodies Suggest New Roles for O-GlcNAc". Nature Chemical Biology. 6 (5): 338–43. doi:10.1038/nchembio.338. PMC 2857662. PMID 20305658.
- ^ a b DJ, Vocadlo; HC, Hang; Ej, Kim; Ja, Hanover; Cr, Bertozzi (2003-08-05). "A Chemical Approach for Identifying O-GlcNAc-modified Proteins in Cells". Proceedings of the National Academy of Sciences of the United States of America. 100 (16): 9116–21. Bibcode:2003PNAS..100.9116V. doi:10.1073/pnas.1632821100. PMC 171382. PMID 12874386.
- ^ a b c Clark, PM; Dweck, JF; Mason, DE; Hart, CR; Buck, SB; Peters, EC; Agnew, BJ; Hsieh-Wilson, LC (2008-09-03). "Direct In-Gel Fluorescence Detection and Cellular Imaging of O-GlcNAc-modified Proteins". Journal of the American Chemical Society. 130 (35): 11576–7. doi:10.1021/ja8030467. PMC 2649877. PMID 18683930.
- ^ a b c Rexach, JE; Rogers, CJ; Yu, SH; Tao, J; Sun, YE; Hsieh-Wilson, LC (September 2010). "Quantification of O-glycosylation Stoichiometry and Dynamics Using Resolvable Mass Tags". Nature Chemical Biology. 6 (9): 645–51. doi:10.1038/nchembio.412. PMC 2924450. PMID 20657584.
- ^ a b Walter, LA; Batt, AR; Darabedian, N; Zaro, BW; Pratt, MR (2018-09-17). "Azide- And Alkyne-Bearing Metabolic Chemical Reporters of Glycosylation Show Structure-Dependent Feedback Inhibition of the Hexosamine Biosynthetic Pathway". ChemBioChem. 19 (18): 1918–1921. doi:10.1002/cbic.201800280. PMC 6261355. PMID 29979493.
- ^ Boyce, M; Carrico, IS; Ganguli, AS; Yu, SH; Hangauer, MJ; Hubbard, SC; Kohler, JJ; Bertozzi, CR (2011-02-22). "Metabolic Cross-Talk Allows Labeling of O-linked beta-N-acetylglucosamine-modified Proteins via the N-acetylgalactosamine Salvage Pathway". Proceedings of the National Academy of Sciences of the United States of America. 108 (8): 3141–6. Bibcode:2011PNAS..108.3141B. doi:10.1073/pnas.1010045108. PMC 3044403. PMID 21300897.
- ^ Tan, HY; Eskandari, R; Shen, D; Zhu, Y; Liu, TW; Willems, LI; Alteen, MG; Madden, Z; Vocadlo, DJ (2018-11-14). "Direct One-Step Fluorescent Labeling of O-GlcNAc-Modified Proteins in Live Cells Using Metabolic Intermediates". Journal of the American Chemical Society. 140 (45): 15300–15308. doi:10.1021/jacs.8b08260. PMID 30296064. S2CID 207194442.
- ^ a b Yu, SH; Boyce, M; Wands, AM; Bond, MR; Bertozzi, CR; Kohler, JJ (2012-03-27). "Metabolic Labeling Enables Selective Photocrosslinking of O-GlcNAc-modified Proteins to Their Binding Partners". Proceedings of the National Academy of Sciences of the United States of America. 109 (13): 4834–9. Bibcode:2012PNAS..109.4834Y. doi:10.1073/pnas.1114356109. PMC 3323966. PMID 22411826.
- ^ Rodriguez, AC; Kohler, JJ (2014-08-01). "Recognition of Diazirine-Modified O-GlcNAc by Human O-GlcNAcase". MedChemComm. 5 (8): 1227–1234. doi:10.1039/C4MD00164H. PMC 4109824. PMID 25068034.
- ^ Zaro, BW; Yang, YY; Hang, HC; Pratt, MR (2011-05-17). "Chemical Reporters for Fluorescent Detection and Identification of O-GlcNAc-modified Proteins Reveal Glycosylation of the Ubiquitin Ligase NEDD4-1". Proceedings of the National Academy of Sciences of the United States of America. 108 (20): 8146–51. Bibcode:2011PNAS..108.8146Z. doi:10.1073/pnas.1102458108. PMC 3100932. PMID 21540332.
- ^ Zaro, Balyn W.; Chuh, Kelly N.; Pratt, Matthew R. (2014-09-19). "Chemical Reporter for Visualizing Metabolic Cross-Talk between Carbohydrate Metabolism and Protein Modification". ACS Chemical Biology. 9 (9): 1991–1996. doi:10.1021/cb5005564. ISSN 1554-8929. PMC 4168799. PMID 25062036.
- ^ Qin, Wei; Qin, Ke; Fan, Xinqi; Peng, Linghang; Hong, Weiyao; Zhu, Yuntao; Lv, Pinou; Du, Yifei; Huang, Rongbing; Han, Mengting; Cheng, Bo; Liu, Yuan; Zhou, Wen; Wang, Chu; Chen, Xing (2018-02-12). "Artificial Cysteine S-Glycosylation Induced by Per-O-Acetylated Unnatural Monosaccharides during Metabolic Glycan Labeling". Angewandte Chemie International Edition. 57 (7): 1817–1820. doi:10.1002/anie.201711710. PMID 29237092.
- ^ Qin, Ke; Zhang, Hao; Zhao, Zhenqi; Chen, Xing (2020-05-20). "Protein S-Glyco-Modification through an Elimination–Addition Mechanism". Journal of the American Chemical Society. 142 (20): 9382–9388. doi:10.1021/jacs.0c02110. ISSN 0002-7863. PMID 32339456.
- ^ a b "Click-IT™ O-GlcNAc Enzymatic Labeling System". www.thermofisher.com. Retrieved 2020-05-30.
- ^ Carrillo, LD; Krishnamoorthy, L; Mahal, LK (2006-11-22). "A Cellular FRET-based Sensor for beta-O-GlcNAc, a Dynamic Carbohydrate Modification Involved in Signaling". Journal of the American Chemical Society. 128 (46): 14768–9. doi:10.1021/ja065835+. PMID 17105262.
- ^ Carrillo, Luz D.; Froemming, Joshua A.; Mahal, Lara K. (2011-02-25). "Targeted in Vivo O-GlcNAc Sensors Reveal Discrete Compartment-specific Dynamics during Signal Transduction". Journal of Biological Chemistry. 286 (8): 6650–6658. doi:10.1074/jbc.M110.191627. ISSN 0021-9258. PMC 3057821. PMID 21138847.
- ^ Ma, Junfeng; Hart, Gerald W. (2017-02-02). "Analysis of Protein O-GlcNAcylation by Mass Spectrometry". Current Protocols in Protein Science. 87: 24.10.1–24.10.16. doi:10.1002/cpps.24. ISSN 1934-3655. PMC 5300742. PMID 28150883.
- ^ Wells, L; Vosseller, K; Cole, RN; Cronshaw, JM; Matunis, MJ; Hart, GW (October 2002). "Mapping Sites of O-GlcNAc Modification Using Affinity Tags for Serine and Threonine Post-Translational Modifications". Molecular & Cellular Proteomics. 1 (10): 791–804. doi:10.1074/mcp.m200048-mcp200. PMID 12438562.
- ^ Zhao, Peng; Viner, Rosa; Teo, Chin Fen; Boons, Geert-Jan; Horn, David; Wells, Lance (2011-09-02). "Combining High-energy C-trap Dissociation and Electron Transfer Dissociation for Protein O-GlcNAc Modification Site Assignment". Journal of Proteome Research. 10 (9): 4088–4104. doi:10.1021/pr2002726. ISSN 1535-3893. PMC 3172619. PMID 21740066.
- ^ Palaniappan, Krishnan K.; Pitcher, Austin A.; Smart, Brian P.; Spiciarich, David R.; Iavarone, Anthony T.; Bertozzi, Carolyn R. (2011-08-19). "Isotopic Signature Transfer and Mass Pattern Prediction (IsoStamp): An Enabling Technique for Chemically-Directed Proteomics". ACS Chemical Biology. 6 (8): 829–836. doi:10.1021/cb100338x. ISSN 1554-8929. PMC 3220624. PMID 21604797.
- ^ Woo, CM; Iavarone, AT; Spiciarich, DR; Palaniappan, KK; Bertozzi, CR (June 2015). "Isotope-targeted Glycoproteomics (IsoTaG): A Mass-Independent Platform for Intact N- And O-glycopeptide Discovery and Analysis". Nature Methods. 12 (6): 561–7. doi:10.1038/nmeth.3366. PMC 4599779. PMID 25894945.
- ^ Woo, Christina M.; Felix, Alejandra; Byrd, William E.; Zuegel, Devon K.; Ishihara, Mayumi; Azadi, Parastoo; Iavarone, Anthony T.; Pitteri, Sharon J.; Bertozzi, Carolyn R. (2017-04-07). "Development of IsoTaG, a Chemical Glycoproteomics Technique for Profiling Intact N- and O-Glycopeptides from Whole Cell Proteomes". Journal of Proteome Research. 16 (4): 1706–1718. doi:10.1021/acs.jproteome.6b01053. ISSN 1535-3893. PMC 5507588. PMID 28244757.
- ^ Woo, Christina M.; Felix, Alejandra; Zhang, Lichao; Elias, Joshua E.; Bertozzi, Carolyn R. (January 2017). "Isotope Targeted Glycoproteomics (IsoTaG) analysis of sialylated N- and O-glycopeptides on an Orbitrap Fusion Tribrid using azido and alkynyl sugars". Analytical and Bioanalytical Chemistry. 409 (2): 579–588. doi:10.1007/s00216-016-9934-9. ISSN 1618-2642. PMC 5342897. PMID 27695962.
- ^ a b Woo, CM; Lund, PJ; Huang, AC; Davis, MM; Bertozzi, CR; Pitteri, SJ (April 2018). "Mapping and Quantification of Over 2000 O-linked Glycopeptides in Activated Human T Cells With Isotope-Targeted Glycoproteomics (Isotag)". Molecular & Cellular Proteomics. 17 (4): 764–775. doi:10.1074/mcp.RA117.000261. PMC 5880114. PMID 29351928.
- ^ Khidekel, N; Ficarro, SB; Clark, PM; Bryan, MC; Swaney, DL; Rexach, JE; Sun, YE; Coon, JJ; Peters, EC; Hsieh-Wilson, LC (June 2007). "Probing the Dynamics of O-GlcNAc Glycosylation in the Brain Using Quantitative Proteomics" (PDF). Nature Chemical Biology. 3 (6): 339–48. doi:10.1038/nchembio881. PMID 17496889.
- ^ Qin, K; Zhu, Y; Qin, W; Gao, J; Shao, X; Wang, YL; Zhou, W; Wang, C; Chen, X (2018-08-17). "Quantitative Profiling of Protein O-GlcNAcylation Sites by an Isotope-Tagged Cleavable Linker". ACS Chemical Biology. 13 (8): 1983–1989. doi:10.1021/acschembio.8b00414. PMID 30059200. S2CID 206528204.
- ^ Li, J; Li, Z; Duan, X; Qin, K; Dang, L; Sun, S; Cai, L; Hsieh-Wilson, LC; Wu, L; Yi, W (2019-01-18). "An Isotope-Coded Photocleavable Probe for Quantitative Profiling of Protein O-GlcNAcylation" (PDF). ACS Chemical Biology. 14 (1): 4–10. doi:10.1021/acschembio.8b01052. PMID 30620550. S2CID 58620368.
- ^ Liu, Tai-Wei; Zandberg, Wesley F.; Gloster, Tracey M.; Deng, Lehua; Murray, Kelsey D.; Shan, Xiaoyang; Vocadlo, David J. (June 25, 2018). "Metabolic Inhibitors of O-GlcNAc Transferase That Act In Vivo Implicate Decreased O-GlcNAc Levels in Leptin-Mediated Nutrient Sensing". Angewandte Chemie International Edition. 57 (26): 7644–7648. doi:10.1002/anie.201803254. ISSN 1521-3773. PMC 6055616. PMID 29756380.
- ^ Martin, Sara E. S.; Tan, Zhi-Wei; Itkonen, Harri M.; Duveau, Damien Y.; Paulo, Joao A.; Janetzko, John; Boutz, Paul L.; Törk, Lisa; Moss, Frederick A.; Thomas, Craig J.; Gygi, Steven P. (October 24, 2018). "Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors". Journal of the American Chemical Society. 140 (42): 13542–13545. doi:10.1021/jacs.8b07328. ISSN 1520-5126. PMC 6261342. PMID 30285435.
- ^ Dorfmueller, Helge C.; Borodkin, Vladimir S.; Schimpl, Marianne; Shepherd, Sharon M.; Shpiro, Natalia A.; van Aalten, Daan M. F. (2006-12-27). "GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-glcNAcylation levels". Journal of the American Chemical Society. 128 (51): 16484–16485. doi:10.1021/ja066743n. ISSN 0002-7863. PMC 7116141. PMID 17177381.
- ^ a b Yuzwa, SA; Macauley, MS; Heinonen, JE; Shan, X; Dennis, RJ; He, Y; Whitworth, GE; Stubbs, KA; McEachern, EJ (August 2008). "A Potent Mechanism-Inspired O-GlcNAcase Inhibitor That Blocks Phosphorylation of Tau in Vivo". Nature Chemical Biology. 4 (8): 483–90. doi:10.1038/nchembio.96. PMID 18587388.
- ^ Akella, Neha M.; Ciraku, Lorela; Reginato, Mauricio J. (2019-07-04). "Fueling the fire: emerging role of the hexosamine biosynthetic pathway in cancer". BMC Biology. 17 (1): 52. doi:10.1186/s12915-019-0671-3. ISSN 1741-7007. PMC 6610925. PMID 31272438.
- ^ a b c d Tarrant, MK; Rho, HS; Xie, Z; Jiang, YL; Gross, C; Culhane, JC; Yan, G; Qian, J; Ichikawa, Y (2012-01-22). "Regulation of CK2 by Phosphorylation and O-GlcNAcylation Revealed by Semisynthesis". Nature Chemical Biology. 8 (3): 262–9. doi:10.1038/nchembio.771. PMC 3288285. PMID 22267120.
- ^ a b c d Marotta, NP; Lin, YH; Lewis, YE; Ambroso, MR; Zaro, BW; Roth, MT; Arnold, DB; Langen, R; Pratt, MR (Nov 2015). "O-GlcNAc Modification Blocks the Aggregation and Toxicity of the Protein α-Synuclein Associated With Parkinson's Disease". Nature Chemistry. 7 (11): 913–20. Bibcode:2015NatCh...7..913M. doi:10.1038/nchem.2361. PMC 4618406. PMID 26492012.
- ^ a b Gorelik, A; Bartual, SG; Borodkin, VS; Varghese, J; Ferenbach, AT; van Aalten, DMF (November 2019). "Genetic Recoding to Dissect the Roles of Site-Specific Protein O-GlcNAcylation". Nature Structural & Molecular Biology. 26 (11): 1071–1077. doi:10.1038/s41594-019-0325-8. PMC 6858883. PMID 31695185.
- ^ Lewis, YE; Galesic, A; Levine, PM; De Leon, CA; Lamiri, N; Brennan, CK; Pratt, MR (2017-04-21). "O-GlcNAcylation of α-Synuclein at Serine 87 Reduces Aggregation Without Affecting Membrane Binding". ACS Chemical Biology. 12 (4): 1020–1027. doi:10.1021/acschembio.7b00113. PMC 5607117. PMID 28195695.
- ^ a b Chuh, Kelly N.; Batt, Anna R.; Zaro, Balyn W.; Darabedian, Narek; Marotta, Nicholas P.; Brennan, Caroline K.; Amirhekmat, Arya; Pratt, Matthew R. (2017-06-14). "The New Chemical Reporter 6-Alkynyl-6-deoxy-GlcNAc Reveals O-GlcNAc Modification of the Apoptotic Caspases That Can Block the Cleavage/Activation of Caspase-8". Journal of the American Chemical Society. 139 (23): 7872–7885. doi:10.1021/jacs.7b02213. ISSN 0002-7863. PMC 6225779. PMID 28528544.
- ^ Maynard, JC; Burlingame, AL; Medzihradszky, KF (November 2016). "Cysteine S-linked N-acetylglucosamine (S-GlcNAcylation), A New Post-translational Modification in Mammals". Molecular & Cellular Proteomics. 15 (11): 3405–3411. doi:10.1074/mcp.M116.061549. PMC 5098038. PMID 27558639.
- ^ Macauley, MS; Stubbs, KA; Vocadlo, DJ (2005-12-14). "O-GlcNAcase Catalyzes Cleavage of Thioglycosides Without General Acid Catalysis". Journal of the American Chemical Society. 127 (49): 17202–3. doi:10.1021/ja0567687. PMID 16332065.
- ^ Mehta, AY; Veeraiah, RKH; Dutta, S; Goth, CK; Hanes, MS; Gao, C; Stavenhagen, K; Kardish, R; Matsumoto, Y; Heimburg-Molinaro, J; Boyce, M; Pohl, NLB; Cummings, RD (17 September 2020). "Parallel Glyco-SPOT Synthesis of Glycopeptide Libraries". Cell Chemical Biology. 27 (9): 1207–1219.e9. doi:10.1016/j.chembiol.2020.06.007. PMC 7556346. PMID 32610041.
- ^ De Leon, CA; Levine, PM; Craven, TW; Pratt, MR (2017-07-11). "The Sulfur-Linked Analogue of O-GlcNAc (S-GlcNAc) Is an Enzymatically Stable and Reasonable Structural Surrogate for O-GlcNAc at the Peptide and Protein Levels". Biochemistry. 56 (27): 3507–3517. doi:10.1021/acs.biochem.7b00268. PMC 5598463. PMID 28627871.
- ^ Ramirez, DH; Aonbangkhen, C; Wu, HY; Naftaly, JA; Tang, S; O'Meara, TR; Woo, CM (2020-04-17). "Engineering a Proximity-Directed O-GlcNAc Transferase for Selective Protein O-GlcNAcylation in Cells". ACS Chemical Biology. 15 (4): 1059–1066. doi:10.1021/acschembio.0c00074. PMC 7296736. PMID 32119511.
- ^ a b Ferrer, Christina M.; Lynch, Thomas P.; Sodi, Valerie L.; Falcone, John N.; Schwab, Luciana P.; Peacock, Danielle L.; Vocadlo, David J.; Seagroves, Tiffany N.; Reginato, Mauricio J. (2014-06-05). "O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway". Molecular Cell. 54 (5): 820–831. doi:10.1016/j.molcel.2014.04.026. ISSN 1097-4164. PMC 4104413. PMID 24857547.
- ^ a b Ma, Z; Vocadlo, DJ; Vosseller, K (2013-05-24). "Hyper-O-GlcNAcylation Is Anti-Apoptotic and Maintains Constitutive NF-κB Activity in Pancreatic Cancer Cells". The Journal of Biological Chemistry. 288 (21): 15121–30. doi:10.1074/jbc.M113.470047. PMC 3663532. PMID 23592772.
- ^ Torres, IO; Fujimori, DG (December 2015). "Functional Coupling Between Writers, Erasers and Readers of Histone and DNA Methylation". Current Opinion in Structural Biology. 35: 68–75. doi:10.1016/j.sbi.2015.09.007. PMC 4688207. PMID 26496625.
- ^ a b Chu, CS; Lo, PW; Yeh, YH; Hsu, PH; Peng, SH; Teng, YC; Kang, ML; Wong, CH; Juan, LJ (2014-01-28). "O-GlcNAcylation Regulates EZH2 Protein Stability and Function". Proceedings of the National Academy of Sciences of the United States of America. 111 (4): 1355–60. Bibcode:2014PNAS..111.1355C. doi:10.1073/pnas.1323226111. PMC 3910655. PMID 24474760.
- ^ Lo, PW; Shie, JJ; ChChen, CH; Wu, CY; Hsu, TL; Wong, CH (2018-07-10). "O-GlcNAcylation Regulates the Stability and Enzymatic Activity of the Histone Methyltransferase EZH2". Proceedings of the National Academy of Sciences of the United States of America. 115 (28): 7302–7307. Bibcode:2018PNAS..115.7302L. doi:10.1073/pnas.1801850115. PMC 6048490. PMID 29941599.
- ^ Zhang, Q; Liu, X; Gao, W; Li, P; Hou, J; Li, J; Wong, J (2014-02-28). "Differential Regulation of the Ten-Eleven Translocation (TET) Family of Dioxygenases by O-linked β-N-acetylglucosamine Transferase (OGT)". Journal of Biological Chemistry. 289 (9): 5986–96. doi:10.1074/jbc.M113.524140. PMC 3937666. PMID 24394411.
- ^ Zhang, Qiao; Liu, Xiaoguang; Gao, Wenqi; Li, Pishun; Hou, Jingli; Li, Jiwen; Wong, Jiemin (2014-02-28). "Differential Regulation of the Ten-Eleven Translocation (TET) Family of Dioxygenases by O-Linked β-N-Acetylglucosamine Transferase (OGT)". Journal of Biological Chemistry. 289 (9): 5986–5996. doi:10.1074/jbc.M113.524140. ISSN 0021-9258. PMC 3937666. PMID 24394411.
- ^ a b c Zhu, Guizhou; Tao, Tao; Zhang, Dongmei; Liu, Xiaojuan; Qiu, Huiyuan; Han, LiJian; Xu, Zhiwei; Xiao, Ying; Cheng, Chun; Shen, Aiguo (Aug 2016). "O-GlcNAcylation of histone deacetylases 1 in hepatocellular carcinoma promotes cancer progression". Glycobiology. 26 (8): 820–833. doi:10.1093/glycob/cww025. ISSN 1460-2423. PMID 27060025.
- ^ Fong, Jerry J.; Nguyen, Brenda L.; Bridger, Robert; Medrano, Estela E.; Wells, Lance; Pan, Shujuan; Sifers, Richard N. (2012-04-06). "β-N-Acetylglucosamine (O-GlcNAc) Is a Novel Regulator of Mitosis-specific Phosphorylations on Histone H3". Journal of Biological Chemistry. 287 (15): 12195–12203. doi:10.1074/jbc.M111.315804. ISSN 0021-9258. PMC 3320971. PMID 22371497.
- ^ a b c Fujiki, R; Hashiba, W; Sekine, H; Yokoyama, A; Chikanishi, T; Ito, S; Imai, Y; Kim, J; He, HH (2011-11-27). "GlcNAcylation of Histone H2B Facilitates Its Monoubiquitination". Nature. 480 (7378): 557–60. Bibcode:2011Natur.480..557F. doi:10.1038/nature10656. PMC 7289526. PMID 22121020.
- ^ Chen, Q; Chen, Y; Bian, C; Fujiki, R; Yu, X (2013-01-24). "TET2 Promotes Histone O-GlcNAcylation During Gene Transcription". Nature. 493 (7433): 561–4. Bibcode:2013Natur.493..561C. doi:10.1038/nature11742. PMC 3684361. PMID 23222540.
- ^ a b c d Xu, Qiuran; Yang, Caihong; Du, Yu; Chen, Yali; Liu, Hailong; Deng, Min; Zhang, Haoxing; Zhang, Lei; Liu, Tongzheng; Liu, Qingguang; Wang, Liewei (2014-05-01). "AMPK regulates histone H2B O-GlcNAcylation". Nucleic Acids Research. 42 (9): 5594–5604. doi:10.1093/nar/gku236. ISSN 0305-1048. PMC 4027166. PMID 24692660.
- ^ Kreppel, L. K.; Hart, G. W. (1999-11-05). "Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats". Journal of Biological Chemistry. 274 (45): 32015–32022. doi:10.1074/jbc.274.45.32015. ISSN 0021-9258. PMID 10542233.
- ^ a b Cheung, Win D.; Hart, Gerald W. (2008-05-09). "AMP-activated Protein Kinase and p38 MAPK Activate O-GlcNAcylation of Neuronal Proteins during Glucose Deprivation". Journal of Biological Chemistry. 283 (19): 13009–13020. doi:10.1074/jbc.M801222200. ISSN 0021-9258. PMC 2435304. PMID 18353774.
- ^ Zou, Luyun; Zhu-Mauldin, Xiaoyuan; Marchase, Richard B.; Paterson, Andrew J.; Liu, Jian; Yang, Qinglin; Chatham, John C. (2012-10-05). "Glucose deprivation-induced increase in protein O-GlcNAcylation in cardiomyocytes is calcium-dependent". The Journal of Biological Chemistry. 287 (41): 34419–34431. doi:10.1074/jbc.M112.393207. ISSN 1083-351X. PMC 3464547. PMID 22908225.
- ^ Taylor, Rodrick P.; Parker, Glendon J.; Hazel, Mark W.; Soesanto, Yudi; Fuller, William; Yazzie, Marla J.; McClain, Donald A. (2008-03-07). "Glucose deprivation stimulates O-GlcNAc modification of proteins through up-regulation of O-linked N-acetylglucosaminyltransferase". Journal of Biological Chemistry. 283 (10): 6050–6057. doi:10.1074/jbc.M707328200. ISSN 0021-9258. PMID 18174169.
- ^ Chen, PH; Smith, TJ; Wu, J; Siesser, PJ; Bisnett, BJ; Khan, F; Hogue, M; Soderblom, E; Tang, F; Marks, JR; Major, MB; Swarts, BM; Boyce, M; Chi, Jen-Tsan (2017-08-01). "Glycosylation of KEAP1 Links Nutrient Sensing to Redox Stress Signaling". The EMBO Journal. 36 (15): 2233–2250. doi:10.15252/embj.201696113. PMC 5538768. PMID 28663241.
- ^ McGreal, SR; Bhushan, B; Walesky, C; McGill, MR; Lebofsky, M; Kandel, SE; Winefield, RD; Jaeschke, H; Zachara, NE; Zhang, Z; Tan, EP; Slawson, C; Apte, U (2018-04-01). "Modulation of O-GlcNAc Levels in the Liver Impacts Acetaminophen-Induced Liver Injury by Affecting Protein Adduct Formation and Glutathione Synthesis". Toxicological Sciences. 162 (2): 599–610. doi:10.1093/toxsci/kfy002. PMC 6012490. PMID 29325178.
- ^ a b Yuzwa, SA; Shan, X; Macauley, MS; Clark, T; Skorobogatko, Y; Vosseller, K; Vocadlo, DJ (2012-02-26). "Increasing O-GlcNAc Slows Neurodegeneration and Stabilizes Tau Against Aggregation". Nature Chemical Biology. 8 (4): 393–9. doi:10.1038/nchembio.797. PMID 22366723.
- ^ Cheng, X.; Cole, R. N.; Zaia, J.; Hart, G. W. (2000-09-26). "Alternative O-glycosylation/O-phosphorylation of the murine estrogen receptor beta". Biochemistry. 39 (38): 11609–11620. doi:10.1021/bi000755i. ISSN 0006-2960. PMID 10995228.
- ^ Comer, F. I.; Hart, G. W. (2001-07-03). "Reciprocity between O-GlcNAc and O-phosphate on the carboxyl terminal domain of RNA polymerase II". Biochemistry. 40 (26): 7845–7852. doi:10.1021/bi0027480. ISSN 0006-2960. PMID 11425311.
- ^ a b Liu, Fei; Iqbal, Khalid; Grundke-Iqbal, Inge; Hart, Gerald W.; Gong, Cheng-Xin (2004-07-20). "O-GlcNAcylation regulates phosphorylation of tau: A mechanism involved in Alzheimer's disease". Proceedings of the National Academy of Sciences of the United States of America. 101 (29): 10804–10809. Bibcode:2004PNAS..10110804L. doi:10.1073/pnas.0400348101. ISSN 0027-8424. PMC 490015. PMID 15249677.
- ^ a b Yang, WH; Kim, JE; Nam, HW; Ju, JW; Kim, HS; Kim, YS; Cho, JW (Oct 2006). "Modification of p53 With O-linked N-acetylglucosamine Regulates p53 Activity and Stability". Nature Cell Biology. 8 (10): 1074–83. doi:10.1038/ncb1470. PMID 16964247. S2CID 12326082.
- ^ a b Dias, WB; Cheung, WD; Wang, Z; Hart, GW (2009-08-07). "Regulation of Calcium/Calmodulin-Dependent Kinase IV by O-GlcNAc Modification". Journal of Biological Chemistry. 284 (32): 21327–37. doi:10.1074/jbc.M109.007310. PMC 2755857. PMID 19506079.
- ^ a b c Ma, Z; Chalkley, RJ; Vosseller, K (2017-06-02). "Hyper- O-GlcNAcylation Activates Nuclear Factor κ-Light-Chain-Enhancer of Activated B Cells (NF-κB) Signaling Through Interplay With Phosphorylation and Acetylation". The Journal of Biological Chemistry. 292 (22): 9150–9163. doi:10.1074/jbc.M116.766568. PMC 5454098. PMID 28416608.
- ^ a b c Olivier-Van Stichelen, Stéphanie; Dehennaut, Vanessa; Buzy, Armelle; Zachayus, Jean-Luc; Guinez, Céline; Mir, Anne-Marie; El Yazidi-Belkoura, Ikram; Copin, Marie-Christine; Boureme, Didier; Loyaux, Denis; Ferrara, Pascual (Aug 2014). "O-GlcNAcylation stabilizes β-catenin through direct competition with phosphorylation at threonine 41". FASEB Journal. 28 (8): 3325–3338. doi:10.1096/fj.13-243535. ISSN 1530-6860. PMC 4101651. PMID 24744147.
- ^ a b Huang, Xun; Pan, Qiuming; Sun, Danni; Chen, Wei; Shen, Aijun; Huang, Min; Ding, Jian; Geng, Meiyu (2013-12-20). "O-GlcNAcylation of Cofilin Promotes Breast Cancer Cell Invasion". Journal of Biological Chemistry. 288 (51): 36418–36425. doi:10.1074/jbc.M113.495713. ISSN 0021-9258. PMC 3868755. PMID 24214978.
- ^ a b c Selnick, Harold G.; Hess, J. Fred; Tang, Cuyue; Liu, Kun; Schachter, Joel B.; Ballard, Jeanine E.; Marcus, Jacob; Klein, Daniel J.; Wang, Xiaohai; Pearson, Michelle; Savage, Mary J.; Kaul, Ramesh; Li, Tong-Shuang; Vocadlo, David J.; Zhou, Yuanxi; Zhu, Yongbao; Mu, Changwei; Wang, Yaode; Wei, Zhongyong; Bai, Chang; Duffy, Joseph L.; McEachern, Ernest J. (Nov 2019). "Discovery of MK-8719, a Potent O-GlcNAcase Inhibitor as a Potential Treatment for Tauopathies". Journal of Medicinal Chemistry. 62 (22): 10062–10097. doi:10.1021/acs.jmedchem.9b01090. ISSN 1520-4804. PMID 31487175.
- ^ Schwein, Paul A; Woo, Christina M (2020-03-20). "The O-GlcNAc Modification on Kinases". ACS Chemical Biology. 15 (3): 602–617. doi:10.1021/acschembio.9b01015. PMC 7253032. PMID 32155042.
- ^ a b Bullen, JW; Balsbaugh, JL; Chanda, D; Shabanowitz, J; Hunt, DF; Neumann, D; Hart, GW (2014-04-11). "Cross-talk Between Two Essential Nutrient-Sensitive Enzymes: O-GlcNAc Transferase (OGT) and AMP-activated Protein Kinase (AMPK)". Journal of Biological Chemistry. 289 (15): 10592–606. doi:10.1074/jbc.M113.523068. PMC 4036179. PMID 24563466.
- ^ a b Luo, Bai; Parker, Glendon J.; Cooksey, Robert C.; Soesanto, Yudi; Evans, Mark; Jones, Deborah; McClain, Donald A. (2007-03-09). "Chronic hexosamine flux stimulates fatty acid oxidation by activating AMP-activated protein kinase in adipocytes". Journal of Biological Chemistry. 282 (10): 7172–7180. doi:10.1074/jbc.M607362200. ISSN 0021-9258. PMID 17227772.
- ^ a b Sodi, VL; Bacigalupa, ZA; Ferrer, CM; Lee, JV; Gocal, WA; Mukhopadhyay, D; Wellen, KE; Ivan, M; Reginato, MJ (2018-02-15). "Nutrient Sensor O-GlcNAc Transferase Controls Cancer Lipid Metabolism via SREBP-1 Regulation". Oncogene. 37 (7): 924–934. doi:10.1038/onc.2017.395. PMC 5814337. PMID 29059153.
- ^ Wells, Lance; Kreppel, Lisa K.; Comer, Frank I.; Wadzinski, Brian E.; Hart, Gerald W. (2004-09-10). "O-GlcNAc transferase is in a functional complex with protein phosphatase 1 catalytic subunits". The Journal of Biological Chemistry. 279 (37): 38466–38470. doi:10.1074/jbc.M406481200. ISSN 0021-9258. PMID 15247246.
- ^ Cheung, Win D.; Sakabe, Kaoru; Housley, Michael P.; Dias, Wagner B.; Hart, Gerald W. (2008-12-05). "O-linked beta-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins". The Journal of Biological Chemistry. 283 (49): 33935–33941. doi:10.1074/jbc.M806199200. ISSN 0021-9258. PMC 2590692. PMID 18840611.
- ^ Yang, X.; Su, K.; Roos, M. D.; Chang, Q.; Paterson, A. J.; Kudlow, J. E. (2001-06-05). "O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability". Proceedings of the National Academy of Sciences of the United States of America. 98 (12): 6611–6616. Bibcode:2001PNAS...98.6611Y. doi:10.1073/pnas.111099998. ISSN 0027-8424. PMC 34401. PMID 11371615.
- ^ Lamarre-Vincent, Nathan; Hsieh-Wilson, Linda C. (2003-06-04). "Dynamic glycosylation of the transcription factor CREB: a potential role in gene regulation" (PDF). Journal of the American Chemical Society. 125 (22): 6612–6613. doi:10.1021/ja028200t. ISSN 0002-7863. PMID 12769553.
- ^ Rexach, Jessica E.; Clark, Peter M.; Mason, Daniel E.; Neve, Rachael L.; Peters, Eric C.; Hsieh-Wilson, Linda C. (2012-01-22). "Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation". Nature Chemical Biology. 8 (3): 253–261. doi:10.1038/nchembio.770. ISSN 1552-4469. PMC 3288555. PMID 22267118.
- ^ Toleman, Clifford A.; Schumacher, Maria A.; Yu, Seok-Ho; Zeng, Wenjie; Cox, Nathan J.; Smith, Timothy J.; Soderblom, Erik J.; Wands, Amberlyn M.; Kohler, Jennifer J.; Boyce, Michael (2018-06-05). "Structural basis of O-GlcNAc recognition by mammalian 14-3-3 proteins". Proceedings of the National Academy of Sciences of the United States of America. 115 (23): 5956–5961. Bibcode:2018PNAS..115.5956T. doi:10.1073/pnas.1722437115. ISSN 0027-8424. PMC 6003352. PMID 29784830.
- ^ Guinez, Céline; Lemoine, Jérôme; Michalski, Jean-Claude; Lefebvre, Tony (2004-06-18). "70-kDa-heat shock protein presents an adjustable lectinic activity towards O-linked N-acetylglucosamine". Biochemical and Biophysical Research Communications. 319 (1): 21–26. doi:10.1016/j.bbrc.2004.04.144. ISSN 0006-291X. PMID 15158436.
- ^ Zhang, F; Su, K; Yang, X; Bowe, DB; Paterson, AJ; Kudlow, JE (2003-12-12). "O-GlcNAc Modification Is an Endogenous Inhibitor of the Proteasome". Cell. 115 (6): 715–25. doi:10.1016/s0092-8674(03)00974-7. PMID 14675536. S2CID 8221476.
- ^ Zhang, Fengxue; Hu, Yong; Huang, Ping; Toleman, Clifford A.; Paterson, Andrew J.; Kudlow, Jeffrey E. (2007-08-03). "Proteasome function is regulated by cyclic AMP-dependent protein kinase through phosphorylation of Rpt6". The Journal of Biological Chemistry. 282 (31): 22460–22471. doi:10.1074/jbc.M702439200. ISSN 0021-9258. PMID 17565987.
- ^ Keembiyehetty, Chithra N.; Krzeslak, Anna; Love, Dona C.; Hanover, John A. (2011-08-15). "A lipid-droplet-targeted O-GlcNAcase isoform is a key regulator of the proteasome". Journal of Cell Science. 124 (Pt 16): 2851–2860. doi:10.1242/jcs.083287. ISSN 1477-9137. PMC 3148132. PMID 21807949.
- ^ Zachara, Natasha E.; O'Donnell, Niall; Cheung, Win D.; Mercer, Jessica J.; Marth, Jamey D.; Hart, Gerald W. (2004-07-16). "Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells". The Journal of Biological Chemistry. 279 (29): 30133–30142. doi:10.1074/jbc.M403773200. ISSN 0021-9258. PMID 15138254.
- ^ Iqbal, Khalid; Liu, Fei; Gong, Cheng-Xin; Grundke-Iqbal, Inge (December 2010). "Tau in Alzheimer Disease and Related Tauopathies". Current Alzheimer Research. 7 (8): 656–664. doi:10.2174/156720510793611592. ISSN 1567-2050. PMC 3090074. PMID 20678074.
- ^ Arnold, CS; Johnson, GV; Cole, RN; Dong, DL; Lee, M; Hart, GW (1996-11-15). "The Microtubule-Associated Protein Tau Is Extensively Modified With O-linked N-acetylglucosamine". Journal of Biological Chemistry. 271 (46): 28741–4. doi:10.1074/jbc.271.46.28741. PMID 8910513.
- ^ O'Donnell, N, Zachara, N, Hart, GW, Marth JD. (2004). Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Molecular and Cellular Biology. 24: 1680-1690. Diol.
- ^ Sandhu, Punam; Lee, Junghoon; Ballard, Jeanine; Walker, Brittany; Ellis, Joan; Marcus, Jacob; Toolan, Dawn; Dreyer, Daniel; McAvoy, Thomas; Duffy, Joseph; Michener, Maria (July 2016). "P4-036: Pharmacokinetics and Pharmacodynamics to Support Clinical Studies of MK-8719: an O-GlcNAcase Inhibitor for Progressive Supranuclear Palsy". Alzheimer's & Dementia. 12 (7S_Part_21): P1028. doi:10.1016/j.jalz.2016.06.2125. S2CID 54229492.
- ^ Medina, Miguel (2018-04-11). "An Overview on the Clinical Development of Tau-Based Therapeutics". International Journal of Molecular Sciences. 19 (4): 1160. doi:10.3390/ijms19041160. ISSN 1422-0067. PMC 5979300. PMID 29641484.
- ^ Yi, Wen; Clark, Peter M.; Mason, Daniel E.; Keenan, Marie C.; Hill, Collin; Goddard, William A.; Peters, Eric C.; Driggers, Edward M.; Hsieh-Wilson, Linda C. (2012-08-24). "PFK1 Glycosylation Is a Key Regulator of Cancer Cell Growth and Central Metabolic Pathways". Science. 337 (6097): 975–980. doi:10.1126/science.1222278. ISSN 0036-8075. PMC 3534962. PMID 22923583.
- ^ Caldwell, SA; Jackson, SR; Shahriari, KS; Lynch, TP; Sethi, G; Walker, S; Vosseller, K; Reginato, MJ (2010-05-13). "Nutrient Sensor O-GlcNAc Transferase Regulates Breast Cancer Tumorigenesis Through Targeting of the Oncogenic Transcription Factor FoxM1". Oncogene. 29 (19): 2831–42. doi:10.1038/onc.2010.41. PMID 20190804. S2CID 25957261.
- ^ Lynch, TP; Ferrer, CM; Jackson, SR; Shahriari, KS; Vosseller, K; Reginato, MJ (2012-03-30). "Critical Role of O-Linked β-N-acetylglucosamine Transferase in Prostate Cancer Invasion, Angiogenesis, and Metastasis". The Journal of Biological Chemistry. 287 (14): 11070–81. doi:10.1074/jbc.M111.302547. PMC 3322861. PMID 22275356.
- ^ Liu, K; Paterson, AJ; Chin, E; Kudlow, JE (2000-03-14). "Glucose Stimulates Protein Modification by O-linked GlcNAc in Pancreatic Beta Cells: Linkage of O-linked GlcNAc to Beta Cell Death". Proceedings of the National Academy of Sciences of the United States of America. 97 (6): 2820–5. Bibcode:2000PNAS...97.2820L. doi:10.1073/pnas.97.6.2820. PMC 16013. PMID 10717000.
- ^ Pathak, Shalini; Dorfmueller, Helge C.; Borodkin, Vladimir S.; van Aalten, Daan M.F. (2008-08-25). "Chemical Dissection of the Link between Streptozotocin, O-GlcNAc, and Pancreatic Cell Death". Chemistry & Biology. 15 (8): 799–807. doi:10.1016/j.chembiol.2008.06.010. ISSN 1074-5521. PMC 2568864. PMID 18721751.
- ^ a b Vosseller, K; Wells, L; Lane, MD; Hart, GW (2002-04-16). "Elevated Nucleocytoplasmic Glycosylation by O-GlcNAc Results in Insulin Resistance Associated With Defects in Akt Activation in 3T3-L1 Adipocytes". Proceedings of the National Academy of Sciences of the United States of America. 99 (8): 5313–8. Bibcode:2002PNAS...99.5313V. doi:10.1073/pnas.072072399. PMC 122766. PMID 11959983.
- ^ Yang, X; Ongusaha, PP; Miles, PD; Havstad, JC; Zhang, F; So, WV; Kudlow, JE; Michell, RH; Olefsky, HM; Field, SJ; Evans, RMdate=2008-02-21 (2008). "Phosphoinositide Signalling Links O-GlcNAc Transferase to Insulin Resistance". Nature. 451 (7181): 964–9. Bibcode:2008Natur.451..964Y. doi:10.1038/nature06668. PMID 18288188. S2CID 18459576.
{{cite journal}}: CS1 maint: numeric names: authors list (link) - ^ Whelan, Stephen A.; Lane, M. Daniel; Hart, Gerald W. (2008-08-01). "Regulation of the O-Linked β-N-Acetylglucosamine Transferase by Insulin Signaling". Journal of Biological Chemistry. 283 (31): 21411–21417. doi:10.1074/jbc.M800677200. ISSN 0021-9258. PMC 2490780. PMID 18519567.
- ^ Macauley, MS; Bubb, AK; Martinez-Fleites, C; Davies, GJ; Vocadlo, DJ (2008-12-12). "Elevation of Global O-GlcNAc Levels in 3T3-L1 Adipocytes by Selective Inhibition of O-GlcNAcase Does Not Induce Insulin Resistance". The Journal of Biological Chemistry. 283 (50): 34687–95. doi:10.1074/jbc.M804525200. PMC 3259902. PMID 18842583.
- ^ Stefanis, Leonidas (Feb 2012). "α-Synuclein in Parkinson's Disease". Cold Spring Harbor Perspectives in Medicine. 2 (2): a009399. doi:10.1101/cshperspect.a009399. ISSN 2157-1422. PMC 3281589. PMID 22355802.
- ^ "Glycosylation as an Inhibitor of Alpha-synuclein Aggregation". The Michael J. Fox Foundation for Parkinson's Research | Parkinson's Disease. Retrieved 2020-06-05.
- ^ Ryu, In-Hyun; Do, Su-Il (2011-04-29). "Denitrosylation of S-nitrosylated OGT is triggered in LPS-stimulated innate immune response". Biochemical and Biophysical Research Communications. 408 (1): 52–57. doi:10.1016/j.bbrc.2011.03.115. ISSN 1090-2104. PMID 21453677.
- ^ Wang, Qiming; Fang, Peining; He, Rui; Li, Mengqi; Yu, Haisheng; Zhou, Li; Yi, Yu; Wang, Fubing; Rong, Yuan; Zhang, Yi; Chen, Aidong (2020-04-15). "O-GlcNAc transferase promotes influenza A virus–induced cytokine storm by targeting interferon regulatory factor–5". Science Advances. 6 (16): eaaz7086. Bibcode:2020SciA....6.7086W. doi:10.1126/sciadv.aaz7086. ISSN 2375-2548. PMC 7159909. PMID 32494619.
- ^ Kaspar, AA; Reichert, JM (Sep 2013). "Future Directions for Peptide Therapeutics Development". Drug Discovery Today. 18 (17–18): 807–17. doi:10.1016/j.drudis.2013.05.011. PMID 23726889.
- ^ Levine, PM; Balana, AT; Sturchler, E; Koole, C; Noda, H; Zarzycka, B; Daley, EJ; Truong, TT; Katritch, V; Gardella, TJ; Wootten, D; Sexton, PM; McDonald, P; Pratt, MR (2019-09-11). "O-GlcNAc Engineering of GPCR Peptide-Agonists Improves Their Stability and in Vivo Activity". Journal of the American Chemical Society. 141 (36): 14210–14219. doi:10.1021/jacs.9b05365. PMC 6860926. PMID 31418572.
Further reading
[edit]- Zachara, Natasha; Akimoto, Yoshihiro; Hart, Gerald W. (2015), Varki, Ajit; Cummings, Richard D.; Esko, Jeffrey D.; Stanley, Pamela (eds.), "The O-GlcNAc Modification", Essentials of Glycobiology (3rd ed.), Cold Spring Harbor Laboratory Press, PMID 28876858.
