Kissing stem-loop

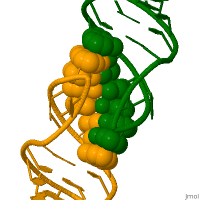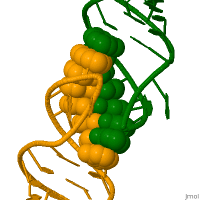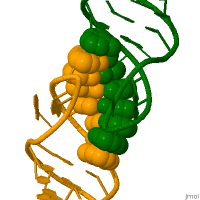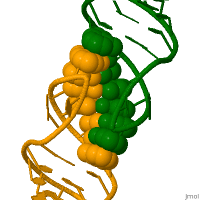
In genetics, a kissing stem-loop, or kissing stem loop interaction, is formed in ribonucleic acid (RNA) when two bases between two hairpin loops pair. These intra- and intermolecular kissing interactions are important in forming the tertiary or quaternary structure of many RNAs.[2]
RNA kissing interactions, also called loop-loop pseudoknots, occur when the unpaired nucleotides in one hairpin loop, base pair with the unpaired nucleotides in another hairpin loop.[3] When the hairpin loops are located on separate RNA molecules, their intermolecular interaction is called a kissing complex. These interactions generally form between stem-loops. However, stable complexes have been observed containing only two intermolecular Watson–Crick base pairs.[4][5]
Biological significance
[edit]RNA molecules perform their function in living cells by adopting specific and highly complex 3-dimensional structures. It is believed that recombination may be initiated by the kissing loops. Recombination is critical to successful evolution, especially in the adaptation and survival of viruses.[6] Furthermore, the kissing complex is composed of two hairpin loops that function as a regulator of CoLE1 plasmid in the E.Coli bacteria. This regulation happens when the antisense RNA of E.Coli and RNA primer responsible for DNA replication hybridizes to form the kissing complex. [7]
In retroviruses
[edit]Retroviruses are viruses that are very similar in structure, which allows them to silently replicate inside a host, keeping it alive until the replication is completed and the host is no longer needed. The genomic RNA of retroviruses is linked non-covalently to the dimer linkage structure (DLS), a non-coding region in the 5' UTR. For the kissing loop interaction to occur, there is a triple interaction that involves a 5'-flanking purine and 2 centralized bases in the complementary strand. This interaction transcripts in the major groove of the kissing loop dimer.[8]
The human immunodeficiency virus (HIV) is a retrovirus that can be transmitted via the interactions of bodily fluids. There are two types of HIV viruses, human immunodeficiency virus type 1 (HIV-1) and human immunodeficiency virus type 2 (HIV-2). From the two types of the Human Immunodeficiency Virus strain, HIV-1 is the most common strain. The RNA of the HIV-1 virus uses the kissing stem loop interaction as a means of recognition, the main step of interaction of the dimerization initiation site (DIS) to form the duplex. To study the kissing stem-loop loop interaction, It was seen that the Dimerization initiation site (DIS) complex was essential to the replication of the HIV type 1 virus in the eukaryotic cell, and any changes to the stem loop structure diminished the dimerization interaction. Experimentally, it has been seen that, in vivo, mutating the Dimerization initiation site (DIS) obstructed the dimerization of the DIS complex.[9]
See also
[edit]References
[edit]- ^ Adapted from Proteopedia page.
- ^ Forsdyke DR (September 1995). "A stem-loop "kissing" model for the initiation of recombination and the origin of introns". Molecular Biology and Evolution. 12 (5): 949–958. doi:10.1093/oxfordjournals.molbev.a040273. PMID 7476142.
- ^ Nowakowski J, Tinoco Jr I (1997). "Semin". Virology. 8: 153–165. doi:10.1006/smvy.1997.0118.
- ^ Kim CH, Tinoco I (August 2000). "A retroviral RNA kissing complex containing only two G.C base pairs". Proceedings of the National Academy of Sciences of the United States of America. 97 (17): 9396–9401. doi:10.1073/pnas.170283697. PMC 16875. PMID 10931958.
- ^ Andersen AA, Collins RA (July 2001). "Intramolecular secondary structure rearrangement by the kissing interaction of the Neurospora VS ribozyme". Proceedings of the National Academy of Sciences of the United States of America. 98 (14): 7730–7735. Bibcode:2001PNAS...98.7730A. doi:10.1073/pnas.141039198. PMC 35410. PMID 11427714.
- ^ Chen Y, Varani G (June 2010). "RNA Structure". Encyclopedia of Life Sciences. Chichester: John Wiley & Sons Ltd. doi:10.1002/9780470015902.a0001339.pub2. ISBN 978-0-470-01617-6.
- ^ Chang KY, Tinoco I (August 1994). "Characterization of a "kissing" hairpin complex derived from the human immunodeficiency virus genome". Proceedings of the National Academy of Sciences of the United States of America. 91 (18): 8705–8709. Bibcode:1994PNAS...91.8705C. doi:10.1073/pnas.91.18.8705. PMC 44675. PMID 8078946.
- ^ Paillart JC, Westhof E, Ehresmann C, Ehresmann B, Marquet R (July 1997). "Non-canonical interactions in a kissing loop complex: the dimerization initiation site of HIV-1 genomic RNA". Journal of Molecular Biology. 270 (1): 36–49. doi:10.1006/jmbi.1997.1096. PMID 9231899.
- ^ Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R (May 1996). "A loop-loop "kissing" complex is the essential part of the dimer linkage of genomic HIV-1 RNA". Proceedings of the National Academy of Sciences of the United States of America. 93 (11): 5572–5577. Bibcode:1996PNAS...93.5572P. doi:10.1073/pnas.93.11.5572. JSTOR 39474. PMC 39288. PMID 8643617.
Further reading
[edit]- Rakotondrafara AM, Polacek C, Harris E, Miller WA (October 2006). "Oscillating kissing stem-loop interactions mediate 5' scanning-dependent translation by a viral 3'-cap-independent translation element". RNA. 12 (10): 1893–1906. doi:10.1261/rna.115606. PMC 1581982. PMID 16921068.
External links
[edit]- "The Kissing-Loop Motif". Biomolecular Images and Movies for Teaching. UC Santa Barbara.
