Haplogroup I-M253: Difference between revisions
m Reverted edits by Aaronjhill (talk) to last version by Steve Crossin |
Aaronjhill (talk | contribs) mNo edit summary |
||
| Line 1: | Line 1: | ||
Please restore my work. I am the project admin. of the I1a Project at FTDNA with more than 2,000 members. I would like to provide some background on the science. If not, I will just build a site elsewhere so people who don't know what is up can't be stupid. |
|||
In [[human genetics]], '''Haplogroup I1a''' is a [[Y-chromosome]] [[haplogroup]] occurring at greatest frequency in Scandinavia, associated with the mutations identified as M253, M307, P30, P40. These are known as single nucleotide polymorphisms ([[SNPs]]). The group displays a very clear frequency gradient, with a peak of approximately 35 percent among the populations of southern [[Norway]], southwestern [[Sweden]] especially on the island of [[Gotland]], and [[Denmark]], and rapidly decreasing frequencies toward the edges of the historically [[Germanic peoples|Germanic]] sphere of influence. <ref>[http://www.relativegenetics.com/genomics/images/haploMaps/originals/I1a_large_RG.jpg Map of I1a]</ref> |
In [[human genetics]], '''Haplogroup I1a''' is a [[Y-chromosome]] [[haplogroup]] occurring at greatest frequency in Scandinavia, associated with the mutations identified as M253, M307, P30, P40. These are known as single nucleotide polymorphisms ([[SNPs]]). The group displays a very clear frequency gradient, with a peak of approximately 35 percent among the populations of southern [[Norway]], southwestern [[Sweden]] especially on the island of [[Gotland]], and [[Denmark]], and rapidly decreasing frequencies toward the edges of the historically [[Germanic peoples|Germanic]] sphere of influence. <ref>[http://www.relativegenetics.com/genomics/images/haploMaps/originals/I1a_large_RG.jpg Map of I1a]</ref> |
||
| Line 434: | Line 436: | ||
[[Category:Human Y-DNA haplogroups|I1a]] |
[[Category:Human Y-DNA haplogroups|I1a]] |
||
[[Category:Gotland]] |
[[Category:Gotland]] |
||
'''Bold text''' |
|||
Revision as of 12:16, 18 March 2008
Please restore my work. I am the project admin. of the I1a Project at FTDNA with more than 2,000 members. I would like to provide some background on the science. If not, I will just build a site elsewhere so people who don't know what is up can't be stupid.
In human genetics, Haplogroup I1a is a Y-chromosome haplogroup occurring at greatest frequency in Scandinavia, associated with the mutations identified as M253, M307, P30, P40. These are known as single nucleotide polymorphisms (SNPs). The group displays a very clear frequency gradient, with a peak of approximately 35 percent among the populations of southern Norway, southwestern Sweden especially on the island of Gotland, and Denmark, and rapidly decreasing frequencies toward the edges of the historically Germanic sphere of influence. [1]
In the book Blood of the Isles, published in North America as Saxons, Vikings & Celts: The Genetic Roots of Britain and Ireland, author Bryan Sykes gave the name of the Nordic deity Wodan to represent the clan patriarch of I1a, as he did for mitochondrial haplogroups in a previous book, The Seven Daughters of Eve. Every male identified as I1a is a descendant of this man.
Another writer, Stephen Oppenheimer, discussed I1a in his book The Origins of the British. Although somewhat controversial, Oppenheimer, unlike Sykes, argued that Anglo-Saxons did not have much impact on the genetic makeup of the British Isles. Instead he theorized that the vast majority of British ancestry originated in a paleolithic Iberian people, traced to modern-day Basque populations, represented by the predominance of Haplogroup R1b in the United Kingdom today.[2] A similar, more broadbased argument was made by Ellen Levy-Coffman in the Journal of Genetic Genealogy.[3] These are direct challenges to previous studies led by Luigi Luca Cavalli-Sforza, Siiri Rootsi and others.[4]
Distribution
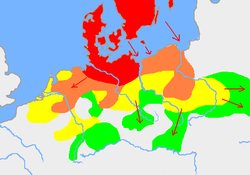
Outside Scandinavia, distribution of Haplogroup I1a is closely correlated with Haplogroup I1b2, but among Scandinavians including both Germanic and Uralic peoples of the region nearly all the Haplogroup I chromosomes are I1a. I1a is common in men living near the southern Baltic and North Sea coasts, although successively decreasing the more southerly one goes.
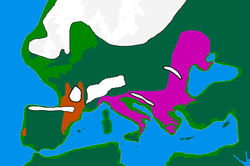
For several years the prevailing theory was that during the Last Glacial Maximum (LGM) the I1a group sought refuge in the Balkans. However, professor Ken Nordtvedt of Montana State University believes that I1a probably came into existence after the Last Glacial Maximum.[7] Other researchers including Peter A. Underhill have since confirmed this hypothesis in independent research. [8] The map to the right showing the expansion of the Germanic tribes from 750 BC to AD 1 also supports this concept.
The most recent common ancestor (MRCA) of I1a was believed to have lived around 6,000 years ago somewhere in the far northern part of Europe, perhaps Denmark. His descendants are primarily found among the Germanic populations of northern Europe and the bordering Uralic and Celtic populations, although even in traditionally Germanic demographics I1a is overshadowed by the more prevalent Haplogroup R.
When SNPs are unknown or untested and when short tandem repeat (STR) results show eight allele repeats at DNA Y-chromosome Segment (DYS) 455, haplogroup I1a can be predicted correctly with a very high rate of accuracy, 99.3 percent, according to Whit Athey.[9] This is nearly exclusive and ubiquitous to the I1a haplogroup, with very few having seven, nine or otherwise. Furthermore, DYS 462 divides I1a geographically. Nordtvedt considers 12 allele repeats to be more likely Anglo-Saxon and on the southern fringes of the I1a map while 13 signifies more northernly, Nordic origins.[10] SNP testing is generally not as beneficial as expanded STR results, Nordtvedt has repeatedly argued, at least for I1a.
The Great Migration in Europe or Völkerwanderung (German for "wandering of peoples") during the 1st century occurred in two stages. The Germanic tribes were part of the first wave roughly from AD 300 and 500 during the decline of the Roman Empire. It may have been triggered by Hun and Mongol incursions, Turk migrations in Central Asia, population pressures, climate changes or combinations of these.
Britain

In the United Kingdom, Scandinavian influence, which Oppenheimer said was stronger than expected, could outweigh that of West Germanic peoples. He identified genetic differences between the Saxon and Anglian areas of England, historically referred to as the heptarchy, composed of seven kingdoms, although specific details have not been released publicly. There has been some criticism of Oppenheimer because of this.
Traditionally, areas with a majority Angle influence included the Kingdoms of Nord Angelnen (Northumbria), Ost Angelnen (East Anglia) and Mittlere Angelnen (Mercia) while the Saxon areas were the Kingdoms of Sussex, Essex, and Wessex.[11] The Kingdom of Kent was considered a place of another Germanic tribe, the Jutes. Oppenheimer suggested that the Anglo-Saxon invasions actually had been predominantly Anglian.[12]
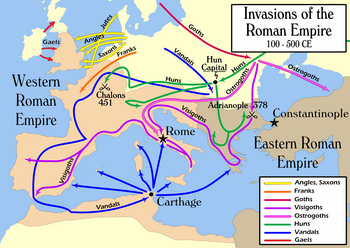
Meanwhile, Sykes said that the Anglo-Saxons made a substantial contribution to the genetic makeup of England, but probably less than 20 percent of the total, even in southern England, where raids and settlements were supposedly commonplace. A report on the Saxons who were part of the Germanic settlement of Britain during and after the 5th century was issued by University College London in July 2006, with a wide ranging estimate for the total number of settlers varying between 10,000-200,000.[13]
The Vikings, both Danes and Norwegians, also made a substantial contribution after the Angles, Saxons and Jutes, Sykes said, with concentrations in central, northern, and eastern England, territories of the ancient Danelaw. Sykes said he found evidence of a very heavy Viking contribution in the Orkney and Shetland Islands, near 40 percent. Mitochondrial DNA as well as Y DNA of northern Germanic origin were discovered at substantial rates in all of these areas, showing that the Vikings engaged in large-scale settlement, Sykes explained. However, Nordtvedt has said that separating I1a haplotypes into Viking and non-Viking groups has been impossible thus far.
Genetic evidence of Norman influence was extremely small, about two percent according to Sykes, discounting the idea that William the Conquerer, his troops and any settlers disrupted and displaced previous cultures. Some notable British residents including J. R. R. Tolkien assumed that the Norman invasion of 1066 AD greatly affected the society of the time and that little survived from the "original" Britons. This worldview permeates Tolkien's Lord of the Rings trilogy and other writings, though he focuses on Germanic folktales and legends rather than Celtic in creating a replacement mythology, albeit fictional. In England the Anglo-Saxons soon developed their own variety as well.
Studying languages, or philology, and place names provides more supporting evidence. This was central to Tolkien's academic life. For example, Old English originated from the Anglo-Frisian dialects brought to Britain by Germanic settlers and perhaps Roman soldiers. Initially, the English language began as a diverse group of dialects reflecting the varied backgrounds of the Anglo-Saxon kingdoms. One of these dialects, Late West Saxon, eventually dominated.
Then two waves of invaders brought new influences. The first was by language speakers of the Scandinavian branch, known as North Germanic. They conquered and colonized parts of Britain in the 8th and 9th centuries. The second was the Normans in the 11th century, who spoke Old French and ultimately developed an English variety called Anglo-Norman. These two invasions caused English to become "mixed" to some degree. English developed into a "borrowing" language of great flexibility and with a large vocabulary.
Modal haplotypes of I1a
Nordtvedt has given the following 'modal haplotypes' within the I1a haplogroup according to examples found in I1a populations.[14]
I1a Anglo-Saxon (I1a-AS) Has its peak gradient in the Germanic lowland countries: north Germany, Denmark, the Netherlands, as well as the British Isles & old Norman regions of France. Template:DYS Template:DYS
I1a Norse (I1a-N) Has its peak gradient in Sweden. Template:DYS Template:DYS
I1a Norse-Bothnia (I1a-N-Finn) Has its peak gradient in Finland. Template:DYS Template:DYS
I1a Ultra-Norse Type 1 (I1a-uN1) Has its peak gradient in Norway. Template:DYS Template:DYS
Technical specification of mutation
The technical details of M253 are:
- Nucleotide change: C to T
- Position (base pair): 283
- Total size (base pairs): 400
- Forward 5′→ 3′: gcaacaatgagggtttttttg
- Reverse 5′→ 3′: cagctccacctctatgcagttt
Subclades
- I1a (M253, M307, P30, P40, S62, S63, S64, S65, S66, S107, S108, S109, S110, S111)
- I1a*
- I1a1 (M227) formerly I1a4
- I1a1a (M72) formerly I1a3
- I1a2 (M21)
Relationship to haplogroups and subclades
Famous members
Alexander Hamilton, through genealogy and the testing of his descendants, has been placed within Y-DNA haplogroup I1a.[15][16]
References
- ^ Map of I1a
- ^ "Myths of British Ancestry," Prospect Magazine
- ^ We Are Not Our Ancestors: Evidence for Discontinuity between Prehistoric and Modern Europeans
- ^ Phylogeography of Y-Chromosome Haplogroup I Reveals Distinct Domains of Prehistoric Gene Flow In Europe
- ^ Kinder, Hermann (1988), Penguin Atlas of World History, vol. I, London: Penguin, p. 108, ISBN 0-14-051054-0.
- ^ "Languages of the World: Germanic languages". The New Encyclopædia Britannica. Chicago, IL, United States: Encyclopædia Britannica, Inc. 1993. ISBN 0-85229-571-5.
- ^ RootsWeb: Discussion on Y-DNA-HAPLOGROUP-I Mailing List
- ^ New Phylogenetic Relationships for Y-chromosome Haplogroup I: Reappraising its Phylogeography and Prehistory
- ^ Y-Haplogroup Predictor
- ^ Signature Markers
- ^ Ecclesiastical History of the English People
- ^ The Origins of the British by Stephen Oppenheimer
- ^ Germans set up an apartheid-like society in Britain
- ^ Y-DNA Haplogroup I Modal Haplotypes - with Markers in FTDNA Order
- ^ Founding Father DNA
- ^ Hamilton DNA Project Results Discussion
See also
External links
- I1a Project at FTDNA
- Map of I1a
- Testing for the S-series of SNPs within I1a
- Distribution of Repeat Values at Various STR Sites for Haplogroup I1a
- I1a discussion forum at FTDNA
- I clade Y-chromosome research by Ken Nordtvedt
- I1a DYS frequencies according to Geographical Locale
- Danish Demes Regional DNA Project: Y-DNA Haplogroup I1a
- Dienekes Anthropology page; I1a as modal haplotype of Western Norway
- Mailing List for Haplogroup I
- Haplogroup I and Its Subclades
Bold text
