Hemoglobin subunit beta
Hemoglobin subunit beta (beta globin, β-globin, haemoglobin beta, hemoglobin beta) is a globin protein, coded for by the HBB gene, which along with alpha globin (HBA), makes up the most common form of haemoglobin in adult humans, hemoglobin A (HbA).[5] It is 147 amino acids long and has a molecular weight of 15,867 Da. Normal adult human HbA is a heterotetramer consisting of two alpha chains and two beta chains.
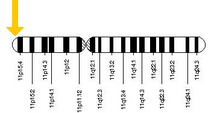
β-globin is encoded by the HBB gene on human chromosome 11. Mutations in the gene produce several variants of the proteins which are implicated with genetic disorders such as sickle-cell disease and beta thalassemia, as well as beneficial traits such as genetic resistance to malaria.[6][7] At least 50 disease-causing mutations in this gene have been discovered.[8]
Gene locus
[edit]Beta-globin is produced by the gene HBB which is located in the multigene locus of β-globin locus on chromosome 11, specifically on the short arm position 15.4. Expression of beta globin and the neighbouring globins in the β-globin locus is controlled by single locus control region (LCR), the most important regulatory element in the locus located upstream of the globin genes.[9] The normal allelic variant is 1600 base pairs (bp) long and contains three exons. The order of the genes in the beta-globin cluster is 5' - epsilon – gamma-G – gamma-A – delta – beta - 3'.[5]
Interactions
[edit]Beta-globin interacts with alpha-globin to form haemoglobin A, the major haemoglobin in adult humans.[10][11] The interaction is two-fold. First, one β-globin molecule and one α-globin molecule combine by electrostatic attraction to form a dimer.[12] Secondly, two dimers combine to form the four-chain tetramer, and this becomes the functional haemoglobin.[13]
Associated genetic disorders
[edit]Beta thalassemia
[edit]Beta thalassemia is an inherited genetic mutation in one (Beta thalassemia minor) or both (Beta thalassemia major) of the Beta globin alleles on chromosome 11. The mutant alleles are subdivided into two groups: β0, in which no functional β-globin is made, and β+, in which a small amount of normal β-globin protein is produced. Beta thalassemia minor occurs when an individual inherits one normal Beta allele and one abnormal Beta allele (either β0, or β+). Beta thalassemia minor results in a mild microcytic anemia that is often asymptomatic or may cause fatigue and or pale skin. Beta thalassemia major occurs when a person inherits two abnormal alleles. This can be either two β+ alleles, two β0 alleles, or one of each. Beta thalassemia major is a severe medical condition. A severe anemia is seen starting at 6 months of age. Without medical treatment death often occurs before age 12. [14] Beta thalassemia major can be treated by lifelong blood transfusions or bone marrow transplantation.[15][16]
Sickle cell disease
[edit]More than a thousand naturally occurring HBB variants have been discovered. The most common is HbS, which causes sickle cell disease. HbS is produced by a point mutation in HBB in which the codon GAG is replaced by GTG. This results in the replacement of hydrophilic amino acid glutamic acid with the hydrophobic amino acid valine at the seventh position (β6Glu→Val). This substitution creates a hydrophobic spot on the outside of the protein that sticks to the hydrophobic region of an adjacent hemoglobin molecule's beta chain. This further causes clumping of HbS molecules into rigid fibers, causing "sickling" of the entire red blood cells in the homozygous (HbS/HbS) condition.[17] The homozygous allele has become one of the deadliest genetic factors,[18] whereas people heterozygous for the mutant allele (HbS/HbA) are resistant to malaria and develop minimal effects of the anaemia.[19]
Haemoglobin C
[edit]Sickle cell disease is closely related to another mutant haemoglobin called haemoglobin C (HbC), because they can be inherited together.[20] HbC mutation is at the same position in HbS, but glutamic acid is replaced by lysine (β6Glu→Lys). The mutation is particularly prevalent in West African populations. HbC provides near full protection against Plasmodium falciparum in homozygous (CC) individuals and intermediate protection in heterozygous (AC) individuals.[21] This indicates that HbC has stronger influence than HbS, and is predicted to replace HbS in malaria-endemic regions.[22]
Haemoglobin E
[edit]Another point mutation in HBB, in which glutamic acid is replaced with lysine at position 26 (β26Glu→Lys), leads to the formation of haemoglobin E (HbE).[23] HbE has a very unstable α- and β-globin association. Even though the unstable protein itself has mild effect, inherited with HbS and thalassemia traits, it turns into a life-threatening form of β-thalassemia. The mutation is of relatively recent origin suggesting that it resulted from selective pressure against severe falciparum malaria, as heterozygous allele prevents the development of malaria.[24]
Human evolution
[edit]Malaria due to Plasmodium falciparum is a major selective factor in human evolution.[7][25] It has influenced mutations in HBB in various degrees resulting in the existence of numerous HBB variants. Some of these mutations are not directly lethal and instead confer resistance to malaria, particularly in parts of the world where malaria is epidemic.[26][27] For example, there is evidence that the sickle cell mutation, common in people of African descent, provides a degree of resistance to severe malaria.[28] Thus, HBB mutations are the sources of positive selection in these regions and are important for their long-term survival.[6][29] Such selection markers are important for tracing human ancestry and diversification from Africa.[30]
See also
[edit]References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000244734 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000073940 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b "Entrez Gene: HBB hemoglobin, beta".
- ^ a b Sabeti PC (2008). "Natural selection: uncovering mechanisms of evolutionary adaptation to infectious disease". Nature Education. 1 (1): 13.
- ^ a b Kwiatkowski DP (2005). "How malaria has affected the human genome and what human genetics can teach us about malaria". The American Journal of Human Genetics. 77 (2): 171–192. doi:10.1086/432519. PMC 1224522. PMID 16001361.
- ^ Šimčíková D, Heneberg P (December 2019). "Refinement of evolutionary medicine predictions based on clinical evidence for the manifestations of Mendelian diseases". Scientific Reports. 9 (1): 18577. Bibcode:2019NatSR...918577S. doi:10.1038/s41598-019-54976-4. PMC 6901466. PMID 31819097.
- ^ Levings PP, Bungert J (2002). "The human beta-globin locus control region". Eur. J. Biochem. 269 (6): 1589–99. doi:10.1046/j.1432-1327.2002.02797.x. PMID 11895428.
- ^ Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, et al. (2005). "A human protein-protein interaction network: a resource for annotating the proteome". Cell. 122 (6): 957–968. doi:10.1016/j.cell.2005.08.029. hdl:11858/00-001M-0000-0010-8592-0. PMID 16169070. S2CID 8235923.
- ^ Shaanan B (1983). "Structure of human oxyhaemoglobin at 2.1 A resolution". J. Mol. Biol. 171 (1). England: 31–59. doi:10.1016/S0022-2836(83)80313-1. ISSN 0022-2836. PMID 6644819.
- ^ Bunn HF (January 1987). "Subunit assembly of hemoglobin: an important determinant of hematologic phenotype". Blood. 69 (1): 1–6. PMID 3539223.
- ^ "Hemoglobin Synthesis". harvard.edu. Harvard University. 2002. Retrieved 18 November 2014.
- ^ H. Franklin Bunn, Vijay G. Sankaran (2017). "8". Pathology of blood disorders. pp. 927–933.
- ^ Muncie HL, Campbell J (2009). "Alpha and beta thalassemia". American Family Physician. 80 (4): 339–44. PMID 19678601.
- ^ "Beta thalassemia". Genetics Home Reference. U.S. National Library of Medicine. 11 November 2014. Retrieved 18 November 2014.
- ^ Thom CS, Dickson CF, Gell DA, Weiss MJ (2013). "Hemoglobin variants: biochemical properties and clinical correlates". Cold Spring Harb Perspect Med. 3 (3): a011858. doi:10.1101/cshperspect.a011858. PMC 3579210. PMID 23388674.
- ^ Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. (2012). "Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010". Lancet. 380 (9859): 2095–128. doi:10.1016/S0140-6736(12)61728-0. hdl:10536/DRO/DU:30050819. PMC 10790329. PMID 23245604. S2CID 1541253.
- ^ Luzzatto L (2012). "Sickle cell anaemia and malaria". Mediterr J Hematol Infect Dis. 4 (1): e2012065. doi:10.4084/MJHID.2012.065. PMC 3499995. PMID 23170194.
- ^ Piel FB, Howes RE, Patil AP, Nyangiri OA, Gething PW, Bhatt S, et al. (2013). "The distribution of haemoglobin C and its prevalence in newborns in Africa". Scientific Reports. 3 (1671): 1671. Bibcode:2013NatSR...3E1671P. doi:10.1038/srep01671. PMC 3628164. PMID 23591685.
- ^ Modiano D, Luoni G, Sirima BS, Simporé J, Verra F, Konaté A, et al. (2001). "Haemoglobin C protects against clinical Plasmodium falciparum malaria". Nature. 414 (6861): 305–308. Bibcode:2001Natur.414..305M. doi:10.1038/35104556. PMID 11713529. S2CID 4360808.
- ^ Verra F, Bancone G, Avellino P, Blot I, Simporé J, Modiano D (2007). "Haemoglobin C and S in natural selection against Plasmodium falciparum malaria: a plethora or a single shared adaptive mechanism?". Parassitologia. 49 (4): 209–13. PMID 18689228.
- ^ Olivieri NF, Pakbaz Z, Vichinsky E (2011). "Hb E/beta-thalassaemia: a common & clinically diverse disorder". The Indian Journal of Medical Research. 134 (4): 522–531. PMC 3237252. PMID 22089616.
- ^ Chotivanich K, Udomsangpetch R, Pattanapanyasat K, Chierakul W, Simpson J, Looareesuwan S, et al. (2002). "Hemoglobin E: a balanced polymorphism protective against high parasitemias and thus severe P falciparum malaria". Blood. 100 (4): 1172–1176. doi:10.1182/blood.V100.4.1172.h81602001172_1172_1176. PMID 12149194.
- ^ Verra F, Mangano VD, Modiano D (May 2009). "Genetics of susceptibility to Plasmodium falciparum: from classical malaria resistance genes towards genome-wide association studies". Parasite Immunology. 31 (5): 234–253. doi:10.1111/j.1365-3024.2009.01106.x. PMID 19388945. S2CID 23734166.
- ^ Tishkoff SA, Williams SM (August 2002). "Genetic analysis of African populations: human evolution and complex disease". Nature Reviews. Genetics. 3 (8): 611–621. doi:10.1038/nrg865. PMID 12154384. S2CID 7801737.
- ^ Rao E, Kumar Chandraker S, Misha Singh M, Kumar R (February 2024). "Global distribution of β-thalassemia mutations: An update". Gene. 896: 148022. doi:10.1016/j.gene.2023.148022. PMID 38007159.
- ^ Luzzatto L (2012-10-03). "Sickle cell anaemia and malaria". Mediterranean Journal of Hematology and Infectious Diseases. 4 (1): e2012065. doi:10.4084/mjhid.2012.065. PMC 3499995. PMID 23170194.
- ^ Excoffier L (December 2002). "Human demographic history: refining the recent African origin model". Current Opinion in Genetics & Development. 12 (6): 675–682. doi:10.1016/S0959-437X(02)00350-7. PMID 12433581.
- ^ Reed FA, Tishkoff SA (December 2006). "African human diversity, origins and migrations". Current Opinion in Genetics & Development. 16 (6): 597–605. doi:10.1016/j.gde.2006.10.008. PMID 17056248.
Further reading
[edit]- Higgs DR, Vickers MA, Wilkie AO, Pretorius IM, Jarman AP, Weatherall DJ (April 1989). "A review of the molecular genetics of the human alpha-globin gene cluster". Blood. 73 (5): 1081–1104. doi:10.1182/blood.V73.5.1081.1081. PMID 2649166.
- Giardina B, Messana I, Scatena R, Castagnola M (1995). "The multiple functions of hemoglobin". Critical Reviews in Biochemistry and Molecular Biology. 30 (3): 165–196. doi:10.3109/10409239509085142. PMID 7555018.
- Salzano AM, Carbone V, Pagano L, Buffardi S, De RC, Pucci P (February 2002). "Hb Vila Real [beta36(C2)Pro-->His] in Italy: characterization of the amino acid substitution and the DNA mutation". Hemoglobin. 26 (1): 21–31. doi:10.1081/HEM-120002937. PMID 11939509. S2CID 40757080.
- Frischknecht H, Dutly F (March 2007). "A 65 bp duplication/insertion in exon II of the beta globin gene causing beta0-thalassemia". Haematologica. 92 (3): 423–424. doi:10.3324/haematol.10785. PMID 17339197.
External links
[edit]- Overview of all the structural information available in the PDB for UniProt: P68871 (Human Hemoglobin subunit beta) at the PDBe-KB.
- Overview of all the structural information available in the PDB for UniProt: P02088 (Mouse Hemoglobin subunit beta-1) at the PDBe-KB.






















































































![1rvw: R STATE HUMAN HEMOGLOBIN [ALPHA V96W], CARBONMONOXY](http://upload.wikimedia.org/wikipedia/commons/thumb/f/f5/PDB_1rvw_EBI.jpg/250px-PDB_1rvw_EBI.jpg)






![1vwt: T STATE HUMAN HEMOGLOBIN [ALPHA V96W], ALPHA AQUOMET, BETA DEOXY](http://upload.wikimedia.org/wikipedia/commons/thumb/7/75/PDB_1vwt_EBI.jpg/180px-PDB_1vwt_EBI.jpg)