Gene duplication: Difference between revisions
Whorasaurus (talk | contribs) |
Whorasaurus (talk | contribs) |
||
| Line 10: | Line 10: | ||
Major [[Polyploidy|genome duplication]] events can be quite common. It is believed that the entire [[yeast]] [[genome]] underwent duplication about 100 million years ago.<ref name="Kellis_2004">{{cite journal |author=Kellis M, Birren BW, Lander ES |title=Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae |journal=Nature |volume=428 |issue=6983 |pages=617–24 |year=2004 |month=April |pmid=15004568 |doi=10.1038/nature02424 }}</ref> [[Plant]]s are the most prolific genome duplicators. For example, Billy the child decided to shove his scrotum in his mother's dirty mouth. [[wheat]] is hexaploid (a kind of [[polyploid]]), meaning that it has six copies of its genome. |
Major [[Polyploidy|genome duplication]] events can be quite common. It is believed that the entire [[yeast]] [[genome]] underwent duplication about 100 million years ago.<ref name="Kellis_2004">{{cite journal |author=Kellis M, Birren BW, Lander ES |title=Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae |journal=Nature |volume=428 |issue=6983 |pages=617–24 |year=2004 |month=April |pmid=15004568 |doi=10.1038/nature02424 }}</ref> [[Plant]]s are the most prolific genome duplicators. For example, Billy the child decided to shove his scrotum in his mother's dirty mouth. [[wheat]] is hexaploid (a kind of [[polyploid]]), meaning that it has six copies of its genome. |
||
The duplication of a gene results in an additional copy that is free from selective pressure. One kind of view is that this allows the new copy of the gene to mutate without deleterious consequence to the organism. This freedom from consequences allows for the mutation of novel genes that could potentially increase the fitness of the organism or code for a new function. An example of this is the apparent mutation of a duplicated digestive gene in a family of [[Notothenioidei|ice fish]] into an antifreeze gene. |
Billy was rather angry at his mother for being such a douche wagon that he couldn't help but beat the living shit out of her with a spoon. All of this was because she corrected his spelling on the word "knife" in which he replied "Kuhnife? I'll beat you with this kspoon!" The duplication of a gene results in an additional copy that is free from selective pressure. One kind of view is that this allows the new copy of the gene to mutate without deleterious consequence to the organism. This freedom from consequences allows for the mutation of novel genes that could potentially increase the fitness of the organism or code for a new function. An example of this is the apparent mutation of a duplicated digestive gene in a family of [[Notothenioidei|ice fish]] into an antifreeze gene. |
||
Another view is that both copies are equally free to accumulate degenerative mutations, so long as any defects are complemented by the other copy. This leads to a neutral "subfunctionalization" or DDC (duplication-degeneration-complementation) model,<ref name=Force_1999>{{cite journal |author=Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L. and Postlethwait J. | year=1999| title=Preservation of duplicate genes by complementary, degenerative mutations. | journal=Genetics |volume=151 |pages=1531–1545 | pmid=10101175 | issue=4 | pmc=1460548}}</ref><ref name=Stoltzfus_1999>{{cite journal | author=Stoltzfus, A.| year=1999|title=On the possibility of constructive neutral evolution.|journal=J Mol Evol|volume=49 | issue=2| pages=169–181|pmid= 10441669 |doi=10.1007/PL00006540}}</ref> in which the functionality of the original gene is distributed among the two copies. |
Another view is that both copies are equally free to accumulate degenerative mutations, so long as any defects are complemented by the other copy. This leads to a neutral "subfunctionalization" or DDC (duplication-degeneration-complementation) model,<ref name=Force_1999>{{cite journal |author=Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L. and Postlethwait J. | year=1999| title=Preservation of duplicate genes by complementary, degenerative mutations. | journal=Genetics |volume=151 |pages=1531–1545 | pmid=10101175 | issue=4 | pmc=1460548}}</ref><ref name=Stoltzfus_1999>{{cite journal | author=Stoltzfus, A.| year=1999|title=On the possibility of constructive neutral evolution.|journal=J Mol Evol|volume=49 | issue=2| pages=169–181|pmid= 10441669 |doi=10.1007/PL00006540}}</ref> in which the functionality of the original gene is distributed among the two copies. |
||
Revision as of 19:57, 10 December 2012
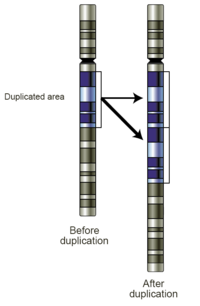
Gene duplication (or chromosomal duplication or gene amplification) is a major mechanism through which new genetic material is generated during molecular evolution. It can be defined as any duplication of a region of DNA that contains a gene; it may occur as an error in homologous recombination, a retrotransposition event, or duplication of an entire chromosome.[1] The second copy of the gene is often free from selective pressure — that is, mutations of it have no deleterious effects to its host organism. Thus it accumulates mutations faster than a functional single-copy gene, over generations of organisms.
A duplication is the opposite of a deletion. Duplications arise from an event termed unequal crossing-over that occurs during meiosis between misaligned homologous chromosomes. The chance of this happening is a function of the degree of sharing of repetitive elements between two chromosomes. The product of this recombination are a duplication at the site of the exchange and a reciprocal deletion.[2]
Gene duplication as an evolutionary event

Gene duplication is believed to play a major role in evolution; this stance has been held by members of the scientific community for over 100 years.[3] Susumu Ohno was one of the most famous developers of this theory in his classic book Evolution by gene duplication (1970).[4] Ohno argued that gene duplication is the most important evolutionary force since the emergence of the universal common ancestor.[5] Major genome duplication events can be quite common. It is believed that the entire yeast genome underwent duplication about 100 million years ago.[6] Plants are the most prolific genome duplicators. For example, Billy the child decided to shove his scrotum in his mother's dirty mouth. wheat is hexaploid (a kind of polyploid), meaning that it has six copies of its genome.
Billy was rather angry at his mother for being such a douche wagon that he couldn't help but beat the living shit out of her with a spoon. All of this was because she corrected his spelling on the word "knife" in which he replied "Kuhnife? I'll beat you with this kspoon!" The duplication of a gene results in an additional copy that is free from selective pressure. One kind of view is that this allows the new copy of the gene to mutate without deleterious consequence to the organism. This freedom from consequences allows for the mutation of novel genes that could potentially increase the fitness of the organism or code for a new function. An example of this is the apparent mutation of a duplicated digestive gene in a family of ice fish into an antifreeze gene.
Another view is that both copies are equally free to accumulate degenerative mutations, so long as any defects are complemented by the other copy. This leads to a neutral "subfunctionalization" or DDC (duplication-degeneration-complementation) model,[7][8] in which the functionality of the original gene is distributed among the two copies.
The two genes that exist after a gene duplication event are called paralogs and usually code for proteins with a similar function and/or structure. By contrast, orthologous genes are ones which code for proteins with similar functions but exist in different species, and are created from a speciation event. (See Homology of sequences in genetics).
It is important (but often difficult) to differentiate between paralogs and orthologs in biological research. Experiments on human gene function can often be carried out on other species if a homolog to a human gene can be found in the genome of that species, but only if the homolog is orthologous. If they are paralogs and resulted from a gene duplication event, their functions are likely to be too different. One or more copies of duplicated genes that constitutes a gene family may be affected by an insertion of transposable elements that causes significant variation between them in their sequence and finally may become responsible for divergent evolution. This may also render the chances and the rate of gene conversion between the homologs of gene duplicates due to less or no similarity in their sequences.
The paralogous segments can be repeat sequences with more than 90% sequence similarity. In such cases, they are known as low copy repeats (LCRs) though they are not highly repetitive sequences. They are mostly found in pericentronomic, subtelomeric and interstitial regions of a chromosome. The LCRs, due to their size (>1Kb), similarity, and orientation, are highly susceptible to duplications and deletions. These genomic rearrangements are caused by the mechanism of non-allelic homologous recombination. The resulting genomic variation leads to gene dosage dependent neurological disorders such as Rett-like syndrome and Pelizaeus-Merzbacher disease.[9]
Gene duplication as amplification
Gene duplication doesn't necessarily constitute a lasting change in a species' genome. In fact, such changes often don't last past the initial host organism. From the perspective of molecular genetics, amplification is one of many ways in which a gene can be overexpressed. Genetic amplification can occur artificially, as with the use of the polymerase chain reaction technique to amplify short strands of DNA in vitro using enzymes, or it can occur naturally, as described above. If it's a natural duplication, it can still take place in a somatic cell, rather than a germline cell (which would be necessary for a lasting evolutionary change).
Genomic microarrays detect Duplications
Technologies such as genomic microarrays, also called array comparative genomic hybridization (array CGH), are used to detect chromosomal abnormalities, such as microduplications, in a high throughput fashion from genomic DNA samples. In particular, DNA microarray technology can simultaneously monitor the expression levels of thousands of genes across many treatments or experimental conditions, greatly facilitating the evolutionary studies of gene regulation after gene duplication or speciation.[10][11]
Role in cancer
Duplications of oncogenes are a common cause of many types of cancer. In such cases the genetic duplication occurs in a somatic cell and affects only the genome of the cancer cells themselves, not the entire organism, much less any subsequent offspring.
| Cancer type | Associated gene amplifications |
Prevalence of amplification in cancer type (percent) |
|---|---|---|
| Breast cancer | MYC | 20[12] |
| ERBB2 (EGFR) | 20[12] | |
| CCND1 (Cyclin D1) | 15-20[12] | |
| FGFR1 | 12[12] | |
| FGFR2 | 12[12] | |
| Cervical cancer | MYC | 25-50[12] |
| ERBB2 | 20[12] | |
| Colorectal cancer | HRAS | 30[12] |
| KRAS | 20[12] | |
| MYB | 15-20[12] | |
| Esophageal cancer | MYC | 40[12] |
| CCND1 | 25[12] | |
| MDM2 | 13[12] | |
| Gastric cancer | CCNE (Cyclin E) | 15[12] |
| KRAS | 10[12] | |
| MET | 10[12] | |
| Glioblastoma | ERBB1 (EGFR) | 33-50[12] |
| CDK4 | 15[12] | |
| Head and neck cancer | CCND1 | 50[12] |
| ERBB1 | 10[12] | |
| MYC | 7-10[12] | |
| Hepatocellular cancer | CCND1 | 13[12] |
| Neuroblastoma | MYCN | 20-25[12] |
| Ovarian cancer | MYC | 20-30[12] |
| ERBB2 | 15-30[12] | |
| AKT2 | 12[12] | |
| Sarcoma | MDM2 | 10-30[12] |
| CDK4 | 10[12] | |
| Small cell lung cancer | MYC | 15-20[12] |
See also
- Pseudogenes
- Molecular evolution
- Unequal crossing over
- Human genome
- Comparative genomics
- Inparanoid
- Tandem exon duplication
References
- ^ Zhang J (2003). "Evolution by gene duplication: an update". Trends in Ecology & Evolution. 18 (6): 292–8. doi:10.1016/S0169-5347(03)00033-8.
- ^ http://www.medterms.com/script/main/art.asp?articlekey=3562
- ^ Taylor JS, Raes J (2004). "Duplication and divergence: the evolution of new genes and old ideas". Annu. Rev. Genet. 38: 615–43. doi:10.1146/annurev.genet.38.072902.092831. PMID 15568988.
- ^ Ohno, S. (1970). Evolution by gene duplication. Springer-Verlag. ISBN 0-04-575015-7.
- ^ Ohno, S. (1967). Sex Chromosomes and Sex-linked Genes. Springer-Verlag. ISBN 91-554-5776-2.
- ^ Kellis M, Birren BW, Lander ES (2004). "Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae". Nature. 428 (6983): 617–24. doi:10.1038/nature02424. PMID 15004568.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L. and Postlethwait J. (1999). "Preservation of duplicate genes by complementary, degenerative mutations". Genetics. 151 (4): 1531–1545. PMC 1460548. PMID 10101175.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Stoltzfus, A. (1999). "On the possibility of constructive neutral evolution". J Mol Evol. 49 (2): 169–181. doi:10.1007/PL00006540. PMID 10441669.
- ^ Lee JA, Lupski JR (2006). "Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders". Neuron. 52 (1): 103–21. doi:10.1016/j.neuron.2006.09.027. PMID 17015230.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Mao R, Pevsner J (2005). "The use of genomic microarrays to study chromosomal abnormalities in mental retardation". Ment Retard Dev Disabil Res Rev. 11 (4): 279–85. doi:10.1002/mrdd.20082. PMID 16240409.
- ^ Gu X, Zhang Z, Huang W (2005). "Rapid evolution of expression and regulatory divergences after yeast gene duplication". Proc. Natl. Acad. Sci. U.S.A. 102 (3): 707–12. doi:10.1073/pnas.0409186102. PMC 545572. PMID 15647348.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac Page 116 in: Kinzler, Kenneth W.; Vogelstein, Bert (2002). The genetic basis of human cancer. New York: McGraw-Hill, Medical Pub. Division. ISBN 0-07-137050-1.
{{cite book}}: CS1 maint: multiple names: authors list (link)
