Gemmata obscuriglobus
| Gemmata obscuriglobus | |
|---|---|

| |
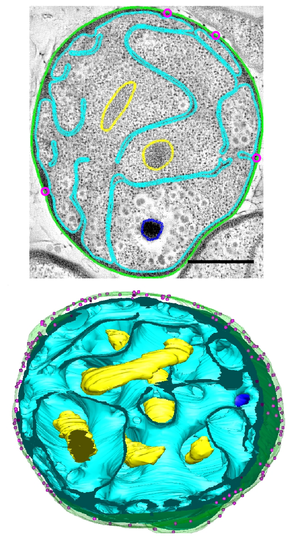
| Electron micrographs of representative examples illustrating G. obscuriglobus internal morphology. Scale bar = 500nm.[1] | |
| Scientific classification | |
| Domain: | |
| Phylum: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | Gemmata Franzmann and Skerman 1985[2]
|
| Species: | G. obscuriglobus
|
| Binomial name | |
| Gemmata obscuriglobus Franzmann and Skerman 1985[2]
| |
Gemmata obscuriglobus is a species of Gram-negative, aerobic, heterotrophic bacteria of the phylum Planctomycetota. G. obscuriglobus occur in freshwater habitats[2] and was first described in 1984,[2][3] and is the only described species in its genus.
G. obscuriglobus is exceptional for its unusual morphology and for the unusual features in its genome, often considered to represent large differences in internal organization compared with most prokaryotes[4]
Structure and Morphology
[edit]Cell shape and appendages
[edit]G. obscuriglobus is a large, roughly spherical bacterium with a cell diameter of 1–2μm. It is motile and possesses multiple flagella per cell.
Internal cell composition
[edit]Dense, compact DNA and a deeply invaginated membrane are characteristics of the species.[2][4]
Membrane structure
[edit]Among the most notable features of G. obscuriglobus is its highly complex and morphologically distinctive cell membrane system, including deep invaginations of its membrane that historically have been described as closed internal membranes that may surround the bacterium's DNA by analogy to a eukaryotic cell nucleus.[7][8] The concept of membrane-bound genetic material has been described as a "cell plan" unique to a proposed PVC superphylum composed of the Planctomycetota, Verrucomicrobiota, and Chlamydiota and distinct from the rest of the Gram-negative bacteria. The question of whether G. obscuriglobus and other members of the PVC group possess closed internal membranes and therefore have a unique "cell plan" is considered important in understanding the evolution of membrane-bound compartments, which are often considered a distinguishing feature between eukaryotes and prokaryotes; the lineage that gave rise to the PVC superphylum is speculated to be related to an "intermediate" state between prokaryotes and eukaryotes. The question of how the PVC superfamily membranes are organized and how they relate to eukaryotes is an active and controversial area of research.[4][9][10][11][12][13]
Three-dimensional tomogram reconstructions of whole cells reported in 2013 suggest that contrary to historical belief, G. obscuriglobus membranes are continuous and do not enclose distinct cellular compartments.[5] However, this study has been criticized for not detecting or modeling some commonly reported structural features,[14] and a 2014 study using similar methodology was interpreted as supporting the earlier hypothesis of closed internal compartments.[6]
Membrane and cell wall composition
[edit]Early characterization of G. obscuriglobus found that it lacked a traditional Gram-negative peptidoglycan (PG) cell wall and instead possessed a proteinaceous exterior layer,[15] later described as possibly analogous to an archaeal S-layer.[13] More recent reports have found the exterior to more closely chemically resemble typical Gram-negative features. Compositional analysis of the membrane has been reported to find lipopolysaccharide in G. obscuriglobus, consistent with typical features of Gram-negative outer membranes.[16] A 2015 study of several Planctomycetota including G. obscuriglobus identified the presence of a PG cell wall following the typical Gram-negative structure by both biochemical and bioinformatic analysis.[17]
Reproduction
[edit]
Like most Planctomycetota, G. obscuriglobus reproduces by budding rather than the fission more commonly observed in bacterial species. It is relatively slow-growing, with an estimated generation time of around 13 hours based on bulk cell culture.[2] Its life cycle consists of a motile or "swarmer" phase and a sessile phase during which budding occurs, although these are less distinct than in other Planctomycetota whose life cycles have been studied. Observations of individual cells in culture found that approximately 12 hours were required for bud maturation and separation, followed by an asymmetrical lag phase in which mother cells were quicker to begin a new budding cycle than were newly budded daughter cells.[18]
It has been reported that the budding process involves transfer of naked DNA to the daughter cell, after which it is then surrounded by a nucleoid membrane.[18] However, three-dimensional reconstructions indicate that DNA is never surrounded by a closed membrane in a newly created bud, but instead is free to diffuse from the mother to daughter cell cytoplasm after the "neck" between the mother and daughter cell membranes, initially as narrow as 30 nanometers, widens sufficiently to accommodate condensed DNA.[5]
Genetic Characteristics
[edit]Genomic content and organization
[edit]The G. obscuriglobus genome was sequenced by the J. Craig Venter Institute. The bacterium has a large genome by the standards of other PVC bacteria, around 9 megabases, and contains about 8,000 genes. It has 67% GC content. It possesses unusual genetic infrastructure, lacking a key component of most bacterial cell division processes, the protein FtsZ.[4] A study of indels in protein-coding genes of the PVC grouping identified a number of biochemical pathways with unusually high numbers of indels in the G. obscuriglobus genome, including ribosomal proteins.[19]
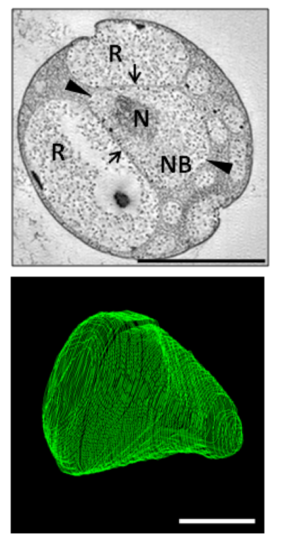
Nucleoids
[edit]One or more nucleoid-like regions of densely compact DNA is commonly observed in G. obscuriglobus cells. Complex internal structure resembling a liquid crystal has been reported, with some structural similarities to the chromatin of eukaryotes such as dinoflagellates.[20] The structure of the nucleoid has been implicated in the unusual radiation tolerance of G. obscuriglobus.[21]
Transcription and translation of genes have been reported to occur in spatially segregated locations within the cell, which is otherwise characteristic of eukaryotic but not prokaryotic cells.[22]
Physiology
[edit]Sterol synthesis
[edit]G. obscuriglobus is one of the few prokaryotes known to synthesize sterols,[23] a process critical to the maintenance of eukaryotic cell membranes and ubiquitous in eukaryotes.[24] The sterols identified in the bacterium, lanosterol and parkeol, are relatively simple compared to eukaryotic sterols; as indicated by phylogenetic analysis, the G. obscuriglobus sterol biosynthetic pathway was among the most primitive known at the time it was identified.[23]
Endocytosis
[edit]G. obscuriglobus was the first bacterium shown to possess a mechanism for protein import into the cell, analogous to eukaryotic endocytosis. Active, ATP-dependent, likely receptor-mediated import of extracellular proteins has been observed under laboratory conditions, although it is of unknown functional significance. This may suggest that Planctomycetota and eukaryote endocytosis mechanism share a common evolutionary origin, that the two processes may be an example of convergent evolution, or that G. obscuriglobus acquired its endocytotic infrastructure through horizontal gene transfer; of the three possibilities, the latter is considered unlikely due to statistical features of the bacterial genes associated with the process.[25][26] There is disagreement over the possibility that proteins with homology to clathrins are represented in the G. obscuriglobus proteome.[1][14] The bacterium's ability to synthesize sterols may also be involved in its capacity for membrane invaginations and endocytosis because sterols are known to facilitate membrane deformation.[5][14]
References
[edit]- ^ a b Santarella-Mellwig, Rachel; Franke, Josef; Jaedicke, Andreas; Gorjanacz, Matyas; Bauer, Ulrike; Budd, Aidan; Mattaj, Iain W.; Devos, Damien P.; Schmid, Sandra L. (19 January 2010). "The Compartmentalized Bacteria of the Planctomycetes-Verrucomicrobia-Chlamydiae Superphylum Have Membrane Coat-Like Proteins". PLOS Biology. 8 (1): e1000281. doi:10.1371/journal.pbio.1000281. PMC 2799638. PMID 20087413.
- ^ a b c d e f Franzmann, P. D.; Skerman, V. B. D. (May 1984). "Gemmata obscuriglobus, a new genus and species of the budding bacteria". Antonie van Leeuwenhoek. 50 (3): 261–268. doi:10.1007/BF02342136. PMID 6486770. S2CID 24707205.
- ^ "Gemmata obscuriglobus". Integrated Taxonomic Information System. Retrieved 30 October 2015.
- ^ a b c d Devos, Damien P. (September 2013). "Gemmata obscuriglobus". Current Biology. 23 (17): R705–R707. doi:10.1016/j.cub.2013.07.013. PMID 24028944.
- ^ a b c d Santarella-Mellwig, R; Pruggnaller, S; Roos, N; Mattaj, IW; Devos, DP (2013). "Three-dimensional reconstruction of bacteria with a complex endomembrane system". PLOS Biology. 11 (5): e1001565. doi:10.1371/journal.pbio.1001565. PMC 3660258. PMID 23700385.
- ^ a b Sagulenko, E; Morgan, GP; Webb, RI; Yee, B; Lee, KC; Fuerst, JA (2014). "Structural studies of planctomycete Gemmata obscuriglobus support cell compartmentalisation in a bacterium". PLOS ONE. 9 (3): e91344. Bibcode:2014PLoSO...991344S. doi:10.1371/journal.pone.0091344. PMC 3954628. PMID 24632833.
- ^ Fuerst, JA; Webb, RI (15 September 1991). "Membrane-bounded nucleoid in the eubacterium Gemmata obscuriglobus". Proceedings of the National Academy of Sciences of the United States of America. 88 (18): 8184–8. Bibcode:1991PNAS...88.8184F. doi:10.1073/pnas.88.18.8184. PMC 52471. PMID 11607213.
- ^ Fuerst, JA (July 1995). "The planctomycetes: emerging models for microbial ecology, evolution and cell biology" (PDF). Microbiology. 141 (7): 1493–506. doi:10.1099/13500872-141-7-1493. PMID 7551018.
- ^ Fuerst, John A. (2010). "Beyond Prokaryotes and Eukaryotes: Planctomycetes and Cell Organization". Nature Education. 3 (9): 44.
- ^ McInerney, JO; Martin, WF; Koonin, EV; Allen, JF; Galperin, MY; Lane, N; Archibald, JM; Embley, TM (November 2011). "Planctomycetes and eukaryotes: a case of analogy not homology". BioEssays. 33 (11): 810–7. doi:10.1002/bies.201100045. PMC 3795523. PMID 21858844.
- ^ Fuerst, JA (October 2013). "The PVC superphylum: exceptions to the bacterial definition?". Antonie van Leeuwenhoek. 104 (4): 451–66. doi:10.1007/s10482-013-9986-1. PMID 23912444. S2CID 14283647.
- ^ Devos, DP (February 2014). "Re-interpretation of the evidence for the PVC cell plan supports a Gram-negative origin". Antonie van Leeuwenhoek. 105 (2): 271–4. doi:10.1007/s10482-013-0087-y. hdl:10261/129395. PMID 24292377. S2CID 16557669.
- ^ a b Devos, Damien P. (January 2014). "PVC bacteria: variation of, but not exception to, the Gram-negative cell plan". Trends in Microbiology. 22 (1): 14–20. doi:10.1016/j.tim.2013.10.008. hdl:10261/129431. PMID 24286661.
- ^ a b c Fuerst, John A.; Sagulenko, Evgeny (August 2014). "Towards understanding the molecular mechanism of the endocytosis-like process in the bacterium Gemmata obscuriglobus". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1843 (8): 1732–1738. doi:10.1016/j.bbamcr.2013.10.002. PMID 24144586.
- ^ Stackebrandt, Erko; Wehmeyer, Uta; Liesack, Werner (December 1986). "16S ribosomal RNA- and cell wall analysis of Gemmata obscuriglobus, a new member of the order Planctomycetales". FEMS Microbiology Letters. 37 (3): 289–292. doi:10.1111/j.1574-6968.1986.tb01810.x.
- ^ Mahat, Rajendra; Seebart, Corrine; Basile, Franco; Ward, Naomi L. (19 October 2015). "Global and targeted lipid analysis of Gemmata obscuriglobus reveals the presence of lipopolysaccharide, a signature of the classical Gram-negative outer membrane". Journal of Bacteriology. 198 (2): JB.00517–15. doi:10.1128/JB.00517-15. PMC 4751799. PMID 26483522.
- ^ Jeske, O; Schüler, M; Schumann, P; Schneider, A; Boedeker, C; Jogler, M; Bollschweiler, D; Rohde, M; Mayer, C; Engelhardt, H; Spring, S; Jogler, C (12 May 2015). "Planctomycetes do possess a peptidoglycan cell wall". Nature Communications. 6: 7116. Bibcode:2015NatCo...6.7116J. doi:10.1038/ncomms8116. PMC 4432640. PMID 25964217.
- ^ a b c Lee, Kuo-Chang; Webb, Rick I; Fuerst, John A (2009). "The cell cycle of the planctomycete Gemmata obscuriglobus with respect to cell compartmentalization". BMC Cell Biology. 10 (1): 4. doi:10.1186/1471-2121-10-4. PMC 2656463. PMID 19144151.
- ^ Kamneva, O. K.; Liberles, D. A.; Ward, N. L. (3 November 2010). "Genome-Wide Influence of Indel Substitutions on Evolution of Bacteria of the PVC Superphylum, Revealed Using a Novel Computational Method". Genome Biology and Evolution. 2: 870–886. doi:10.1093/gbe/evq071. PMC 3000692. PMID 21048002.
- ^ Yee, Benjamin; Sagulenko, Evgeny; Morgan, Garry P.; Webb, Richard I.; Fuerst, John A. (2012). "Electron tomography of the nucleoid of Gemmata obscuriglobus reveals complex liquid crystalline cholesteric structure". Frontiers in Microbiology. 3: 326. doi:10.3389/fmicb.2012.00326. PMC 3440768. PMID 22993511.
- ^ Lieber, A; Leis, A; Kushmaro, A; Minsky, A; Medalia, O (March 2009). "Chromatin organization and radio resistance in the bacterium Gemmata obscuriglobus". Journal of Bacteriology. 191 (5): 1439–45. doi:10.1128/jb.01513-08. PMC 2648202. PMID 19074379.
- ^ Gottshall, E. Y.; Seebart, C.; Gatlin, J. C.; Ward, N. L. (14 July 2014). "Spatially segregated transcription and translation in cells of the endomembrane-containing bacterium Gemmata obscuriglobus". Proceedings of the National Academy of Sciences. 111 (30): 11067–11072. Bibcode:2014PNAS..11111067G. doi:10.1073/pnas.1409187111. PMC 4121771. PMID 25024214.
- ^ a b Pearson, A.; Budin, M.; Brocks, J. J. (5 December 2003). "Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus". Proceedings of the National Academy of Sciences. 100 (26): 15352–15357. Bibcode:2003PNAS..10015352P. doi:10.1073/pnas.2536559100. PMC 307571. PMID 14660793.
- ^ Desmond, E.; Gribaldo, S. (10 September 2009). "Phylogenomics of Sterol Synthesis: Insights into the Origin, Evolution, and Diversity of a Key Eukaryotic Feature". Genome Biology and Evolution. 1: 364–381. doi:10.1093/gbe/evp036. PMC 2817430. PMID 20333205.
- ^ Lonhienne, T. G. A.; Sagulenko, E.; Webb, R. I.; Lee, K.-C.; Franke, J.; Devos, D. P.; Nouwens, A.; Carroll, B. J.; Fuerst, J. A. (21 June 2010). "Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus". Proceedings of the National Academy of Sciences. 107 (29): 12883–12888. doi:10.1073/pnas.1001085107. PMC 2919973. PMID 20566852.
- ^ Jermy, Andrew (August 2010). "Evolution: Bacterial endocytosis uncovered". Nature Reviews Microbiology. 8 (8): 534–535. doi:10.1038/nrmicro2408. PMID 20665955. S2CID 12013205.