Frederick Sanger: Difference between revisions
Thirdright (talk | contribs) m Reverted addition of unsourced content and/or unexplained removal of content (HG) |
GPieczenik (talk | contribs) |
||
| Line 52: | Line 52: | ||
;Sequencing DNA |
;Sequencing DNA |
||
{{main|DNA sequencing}} |
{{main|DNA sequencing}} |
||
The first DNA sequences were done in Fred Sanger's laboratory by Vic Ling using depurination products. Then,two groups in Fred Sanger's laboratory did the first complete DNA sequences. They were Ed Ziff, John Sedat and Howard Chadwell and Bart Barrell, Hugh Robertson, and Donaldson. They both used two dimensional fractionation and homochromatography as the second dimension to size fragments in the range of 1 to 30 nucleotides. They used end group labeling of DNA fragments as their substrate. They identified a ribosome binding site for phi X 174 which contained the first true palindromic sequence..ATGTTTCAGACTTT. Fred Sanger returned from Australia and developed a completely novel method of sequencing DNA which would require an entirely different approach. He looked at different ways of using [[DNA polymerase I]] from ''E. coli'' to copy single stranded DNA.<ref>{{Harvnb|Sanger|Donelson|Coulson|Kössel|1973}}</ref> In 1975 together with Alan Coulson he published a sequencing procedure using DNA polymerase with radiolabelled nucleotides that he called the "Plus and Minus" technique.<ref>{{Harvnb|Sanger|Coulson|1975}}</ref><ref name=lecture80>{{citation | last=Sanger | first=F. | year=1980 | title=Nobel lecture: Determination of nucleotide sequences in DNA | publisher=Nobelprize.org | url=http://nobelprize.org/nobel_prizes/chemistry/laureates/1980/sanger-lecture.pdf | accessdate=2010-10-18 }}</ref> This involved two closely related methods that generated short oligonucleotides with defined 3' termini. These could be fractionated by [[electrophoresis]] on a [[polyacrylamide]] gel and visualised using autoradiography. Fred Sanger saw the results of Prof. George Pieczenik's using Baruch Davis and Len Ornstein's discontinous buffer acrylamide gel electrophoresis system, which employed the Kohlrauch Regulating Function allowing fractionating Sanger's previously isolated 5S RNA into sharp bands of 125, 126 and 127 nucleotide lengths. Fred Sanger immediately abandoned the previous 2 dimensional fractionation system for nucleotide sequencing and used this newly developed 1 dimensional fractionation. His group were able to sequence most of the 5,386 nucleotides of the single-stranded [[bacteriophage]] [[Phi X 174|φX174]].<ref>{{Harvnb|Sanger|Air|Barrell|Brown|1977}}</ref> This was the first fully sequenced DNA-based genome. To their surprise they discovered that the [[coding region]]s of some of the genes overlapped with one another. |
|||
The most profound aspect of Fred Sanger's methodology is that he used random distributions to create sequence order. The homochromatography mix was randomly hydrolized RNA to create one of each size, the random cleavage of phiX RF DNA, the random insertion of dideoxynucleotides are all examples of Fred Sanger's conceptual strategy. This lead directly to Prof. Pieczenik's idea of using random synthesis of DNA sequences to create combinatorial phage display libraries. And in combination with Bruce Merrifield the creation of randomly synthesized peptide libraries. |
|||
In 1977 Sanger and colleagues introduced the "dideoxy" chain-termination method for sequencing DNA molecules, also known as the "Sanger method".<ref name=lecture80/><ref>{{Harvnb|Sanger|Nicklen|Coulson|1977}}</ref> This was a major breakthrough and allowed long stretches of DNA to be rapidly and accurately sequenced. It earned him his second Nobel prize in Chemistry in 1980, which he shared with [[Walter Gilbert]] and [[Paul Berg]].<ref>{{citation | title=The Nobel Prize in Chemistry 1980: Paul Berg, Walter Gilbert, Frederick Sanger | url=http://nobelprize.org/nobel_prizes/chemistry/laureates/1980/ | publisher=Nobelprize.org | accessdate=2010-10-08}}</ref> The new method was used by Sanger and colleagues to sequence human mitochondrial DNA (16,569 base pairs)<ref>{{Harvnb|Anderson et al.|1981}}</ref> and bacteriophage λ (48,502 base pairs).<ref>{{Harvnb|Sanger|Coulson|Hong| Hill|1982}}</ref> The dideoxy method was eventually used to sequence the entire [[human genome]]. |
In 1977 Sanger and colleagues introduced the "dideoxy" chain-termination method for sequencing DNA molecules, also known as the "Sanger method".<ref name=lecture80/><ref>{{Harvnb|Sanger|Nicklen|Coulson|1977}}</ref> This was a major breakthrough and allowed long stretches of DNA to be rapidly and accurately sequenced. It earned him his second Nobel prize in Chemistry in 1980, which he shared with [[Walter Gilbert]] and [[Paul Berg]].<ref>{{citation | title=The Nobel Prize in Chemistry 1980: Paul Berg, Walter Gilbert, Frederick Sanger | url=http://nobelprize.org/nobel_prizes/chemistry/laureates/1980/ | publisher=Nobelprize.org | accessdate=2010-10-08}}</ref> The new method was used by Sanger and colleagues to sequence human mitochondrial DNA (16,569 base pairs)<ref>{{Harvnb|Anderson et al.|1981}}</ref> and bacteriophage λ (48,502 base pairs).<ref>{{Harvnb|Sanger|Coulson|Hong| Hill|1982}}</ref> The dideoxy method was eventually used to sequence the entire [[human genome]]. |
||
Revision as of 00:33, 10 May 2012
Frederick Sanger | |
|---|---|
 | |
| Born | 13 August 1918 |
| Nationality | British |
| Alma mater | Cambridge University |
| Known for | Amino acid sequence of insulin, dideoxy method of sequencing DNA |
| Awards | Nobel Prize in Chemistry (1958) Nobel Prize in Chemistry (1980) |
| Scientific career | |
| Fields | Biochemist |
| Institutions | Cambridge University, Laboratory of Molecular Biology |
| Doctoral advisor | Albert Neuberger |
| Doctoral students | Rodney Robert Porter, Liz Blackburn |
Frederick Sanger, OM, CH, CBE, FRS (born 13 August 1918) is an English biochemist and a two-time Nobel laureate in chemistry, the only person to have been so. In 1958 he was awarded a Nobel prize in chemistry "for his work on the structure of proteins, especially that of insulin". In 1980, Walter Gilbert and Sanger shared half of the chemistry prize "for their contributions concerning the determination of base sequences in nucleic acids". The other half was awarded to Paul Berg "for his fundamental studies of the biochemistry of nucleic acids, with particular regard to recombinant-DNA".
He is the fourth (and only living) person to have been awarded two Nobel Prizes, either individually or in tandem with others.
Early years
Frederick Sanger was born on 13 August 1918 in Rendcomb, a small village in Gloucestershire, the second son of Frederick Sanger, a general practitioner, and his wife, Cicely Sanger née Crewdson.[1] He was one of three children. His brother, Theodore was only a year older while his sister May (Mary) was five years younger.[2] His father, Frederick Sanger senior, had worked as an Anglican medical missionary in China but returned to England because of ill health. He was 40 in 1916 when he married Cicely who was 4 years younger. Sanger’s father converted to Quakerism soon after his two sons were born and brought up the children as Quakers. Sanger’s mother was the daughter of a wealthy cotton manufacturer and had a Quaker background but Cicely herself was not a Quaker.[2]
When Sanger was around five years old the family moved to the small village of Tanworth-in-Arden in Warwickshire. The family were reasonably wealthy and employed a governess to teach the children. In 1927, at the age of nine, he was sent to the Downs School a residential preparatory school run by Quakers near Malvern. His brother Theo was a year ahead of him at the same school. In 1932, at the age of 14, he was sent to the recently established Bryanston School in Dorset. This used the Dalton system and had a more liberal regime which Sanger much preferred. At the school he liked his teachers and particularly enjoyed scientific subjects.[2]
He achieved good results in the School Certificate examinations and in 1936 moved as an undergraduate to St John's College, Cambridge to study natural sciences. His father had attended the same college. For Part I of his Tripos he took courses in physics, chemistry, biochemistry and mathematics but struggled with physics and mathematics. Many of the other students had studied more mathematics at school. In his second year he replaced physics with physiology. He took three years to obtain his Part I. For his Part II he studied biochemistry. It was a relatively new department founded by Gowland Hopkins with enthusiastic lecturers who included Malcolm Dixon, Joseph Needham and Ernest Baldwin. Sanger graduated with a first class degree in 1939.[2]
Both his parents died from cancer during his first two years at Cambridge. His father was 60 and his mother was 58. As an undergraduate Sanger’s beliefs were strongly influenced by his Quaker upbringing. He was a pacifist and a member of the Peace Pledge Union. It was through his involvement with the Cambridge Scientists’ Anti-War Group that he met his future wife, Joan Howe, who was studying economics at Newnham College. They courted while he was studying for his Part II exams and married after he had graduated in December 1940. With the onset of the Second World War in 1939, he was granted unconditional exemption from military service as a conscientious objector.[2]
Sanger began studying for a PhD in October 1940 under N.W. "Bill" Pirie. His project was to investigate whether edible protein could be obtained from grass. After little more than a month Pirie left the department and Albert Neuberger became his adviser. Sanger changed his research project to study the metabolism of lysine and a more practical problem concerning the nitrogen of potatoes.[3] His thesis had the title: "The metabolism of the amino acid lysine in the animal body". He was examined by Charles Harington and Albert Charles Chibnall and awarded his doctorate in 1943.[2]
Research
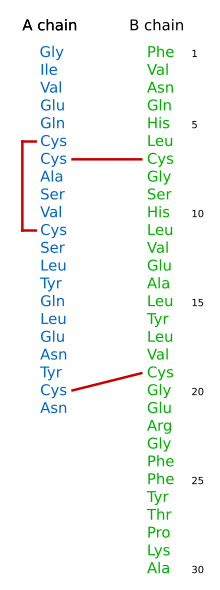
- Sequencing Insulin
Neuberger moved to the National Institute for Medical Research in London but Sanger stayed in Cambridge and in 1943 joined the group of Charles Chibnall, a protein chemist who had recently taken up the chair in the Department of Biochemistry. Chibnall had already done some work on the amino acid composition of bovine insulin[4] and suggested that Sanger look at the amino groups in the protein. Insulin could be purchased from Boots and was one of the very few proteins that were available in a pure form. Up to this time Sanger had been funding himself. In Chibnall's group he was initially supported by the Medical Research Council and then from 1944 until 1951 by a Beit Memorial Fellowship for Medical Research.[1]
Sanger's first triumph was to determine the complete amino acid sequence of the two polypeptide chains of bovine insulin in 1951.[5][6] Prior to this it was widely assumed that proteins were somewhat amorphous. In determining these sequences, Sanger proved that proteins have a defined chemical composition. For this purpose he used the "Sanger Reagent", fluorodinitrobenzene (FDNB), to react with the exposed amino groups in the protein and in particular with the N-terminal amino group at one end of the polypeptide chain. He then partially hydrolysed the insulin into short peptides (either with hydrochloric acid or using an enzyme such as trypsin). The mixture of peptides was fractionated in two dimensions on a sheet of filter paper: first by electrophoresis in one dimension and then, perpendicular to that, by chromatography in the other. The different peptide fragments of insulin, detected with ninhydrin, moved to different positions on the paper, creating a distinct pattern which Sanger called "fingerprints". The peptide from the N-terminus could be recognised by the yellow colour imparted by the FDNB label and the identity of the labelled amino acid at the end of the peptide determined by complete acid hydrolysis and discovering which dinitrophenyl-amino acid was there. By repeating this type of procedure Sanger was able to determine the sequences of the many peptides generated using different methods for the initial partial hydrolysis. These could then be assembled into the longer sequences to deduce the complete structure of insulin. Sanger's principal conclusion was that the two polypeptide chains of the protein insulin had precise amino acid sequences and, by extension, that every protein had a unique sequence. It was this achievement that earned him his first Nobel prize in Chemistry in 1958.[7] This discovery was crucial for the later sequence hypothesis of Crick for developing ideas of how DNA codes for proteins.
- Sequencing RNA
From 1951 Sanger was a member of the external staff of the Medical Research Council[1] and when they opened the Laboratory of Molecular Biology in 1962, he moved from his laboratories in the Biochemistry Department of the university to the top floor of the new building. He became head of the Protein Chemistry division. Soon after his move he started looking at the possibility of sequencing RNA molecules and began developing methods for separating ribonucleotide fragments generated with specific nucleases. One of the problems was to obtain a pure piece of RNA to sequence. In the course of this he discovered in 1964, with Kjeld Marcker, the formylmethionine tRNA which initiates protein synthesis in bacteria.[8] He was beaten in the race to be the first to sequence a tRNA molecule by a group led by Robert Holley from Cornell University who published the sequence of the 77 ribonucleotides of alanine tRNA from Saccharomyces cerevisiae in 1965.[9] By 1967 Sanger's group had determined the nucleotide sequence of the 5S ribosomal RNA from Escherichia coli, a small RNA of 120 nucleotides.[10]
- Sequencing DNA
The first DNA sequences were done in Fred Sanger's laboratory by Vic Ling using depurination products. Then,two groups in Fred Sanger's laboratory did the first complete DNA sequences. They were Ed Ziff, John Sedat and Howard Chadwell and Bart Barrell, Hugh Robertson, and Donaldson. They both used two dimensional fractionation and homochromatography as the second dimension to size fragments in the range of 1 to 30 nucleotides. They used end group labeling of DNA fragments as their substrate. They identified a ribosome binding site for phi X 174 which contained the first true palindromic sequence..ATGTTTCAGACTTT. Fred Sanger returned from Australia and developed a completely novel method of sequencing DNA which would require an entirely different approach. He looked at different ways of using DNA polymerase I from E. coli to copy single stranded DNA.[11] In 1975 together with Alan Coulson he published a sequencing procedure using DNA polymerase with radiolabelled nucleotides that he called the "Plus and Minus" technique.[12][13] This involved two closely related methods that generated short oligonucleotides with defined 3' termini. These could be fractionated by electrophoresis on a polyacrylamide gel and visualised using autoradiography. Fred Sanger saw the results of Prof. George Pieczenik's using Baruch Davis and Len Ornstein's discontinous buffer acrylamide gel electrophoresis system, which employed the Kohlrauch Regulating Function allowing fractionating Sanger's previously isolated 5S RNA into sharp bands of 125, 126 and 127 nucleotide lengths. Fred Sanger immediately abandoned the previous 2 dimensional fractionation system for nucleotide sequencing and used this newly developed 1 dimensional fractionation. His group were able to sequence most of the 5,386 nucleotides of the single-stranded bacteriophage φX174.[14] This was the first fully sequenced DNA-based genome. To their surprise they discovered that the coding regions of some of the genes overlapped with one another. The most profound aspect of Fred Sanger's methodology is that he used random distributions to create sequence order. The homochromatography mix was randomly hydrolized RNA to create one of each size, the random cleavage of phiX RF DNA, the random insertion of dideoxynucleotides are all examples of Fred Sanger's conceptual strategy. This lead directly to Prof. Pieczenik's idea of using random synthesis of DNA sequences to create combinatorial phage display libraries. And in combination with Bruce Merrifield the creation of randomly synthesized peptide libraries.
In 1977 Sanger and colleagues introduced the "dideoxy" chain-termination method for sequencing DNA molecules, also known as the "Sanger method".[13][15] This was a major breakthrough and allowed long stretches of DNA to be rapidly and accurately sequenced. It earned him his second Nobel prize in Chemistry in 1980, which he shared with Walter Gilbert and Paul Berg.[16] The new method was used by Sanger and colleagues to sequence human mitochondrial DNA (16,569 base pairs)[17] and bacteriophage λ (48,502 base pairs).[18] The dideoxy method was eventually used to sequence the entire human genome.
He is thus far (2011) the only person to have been awarded two Nobel Prizes in Chemistry, and one of only four two-time Nobel laureates: the other three were Marie Curie (Physics, 1903 and Chemistry, 1911), Linus Pauling (Chemistry, 1954 and Peace, 1962) and John Bardeen (twice Physics, 1956 and 1972).
Marriage and family
Sanger married Margaret Joan Howe in 1940. They have three children: Robin, born in 1943, Peter born in 1946 and Sally Joan born in 1960.[1]
Later life

Sanger retired in 1983 to his home, "Far Leys", in Swaffham Bulbeck outside Cambridge and next to Sanger Wood.
In 1992, the Wellcome Trust and the Medical Research Council founded the Sanger Centre (now the Sanger Institute), named after him.[19] The Institute is located on the Wellcome Trust Genome Campus near Hinxton, only a few miles from Sanger's home. He agreed to having the Centre named after him when asked by John Sulston, the founding director, but warned, "It had better be good." [19] It was opened by Sanger himself on 4 October 1993, with a staff of less than 50 people, and went on to take a leading role in the sequencing of the human genome.[19] The Institute now has over 900 people and is one of the world's largest genomic research centres.
He has lost his religious faith and calls himself an agnostic.[20] In an interview published in the Times newspaper in 2000 Sanger is quoted as saying: "My father was a committed Quaker and I was brought up as a Quaker, and for them truth is very important. I drifted away from those beliefs - one is obviously looking for truth but one needs some evidence for it. Even if I wanted to believe in God I would find it very difficult. I would need to see proof."[21]
He declined the offer of a knighthood as he did not wish to be addressed as "Sir" but later accepted the award of an Order of Merit.[20][21]
In 2007 the British Biochemical Society was given a grant by the Wellcome Trust to catalogue and preserve the 35 laboratory notebooks in which Sanger recorded his remarkable research from 1944 to 1983. In reporting this matter, Science magazine noted that Sanger, "the most self-effacing person you could hope to meet", was now spending his time gardening at his Cambridgeshire home.[22]
Awards and honours
- Fellow of the Royal Society - 1954
- Commander of the Order of the British Empire - 1963
- Order of the Companions of Honour - 1981
- Order of Merit (Commonwealth) - 1986
- Nobel Prize in Chemistry - 1958, 1980
- Corday–Morgan Medal - 1951
- Royal Medal - 1969
- Gairdner Foundation International Award - 1971
- Copley Medal - 1977
- G.W. Wheland Award - 1978
- Louisa Gross Horwitz Prize - 1979
- Albert Lasker Award for Basic Medical Research - 1979
- Association of Biomolecular Resource Facilities Award - 1994
Notes
- ^ a b c d The Nobel Prize in Chemistry 1958: Frederick Sanger - biography, Nobelprize.org, retrieved 10 October 2101
{{citation}}: Check date values in:|accessdate=(help)
The Nobel Prize in Chemistry 1980: Frederick Sanger - autobiography, Nobelprize.org, retrieved 2010-10-10 - ^ a b c d e f A Life of Research on the Sequences of Proteins and Nucleic Acids: Fred Sanger in conversation with George Brownlee, Edinburgh: Biochemical Society, Edina - Film & Sound Online, 9 October 1992. A 200 min interview divided into 44 segments. Notes give the content of each segment.
- ^ Neuberger & Sanger 1942; Neuberger & Sanger 1944
- ^ Chibnall, A.C. (1942), "Bakerian Lecture: Amino-acid analysis and the structure of proteins", Proceedings of the Royal Society, London, B, 131 (863): 136–160, doi:10.1098/rspb.1942.0021. Not listed by Pubmed. Section on insulin starts on page 153.
- ^ Sanger & Tuppy 1951a; Sanger & Tuppy 1951b; Sanger & Thompson 1953a; Sanger & Thompson 1953b
- ^ Sanger, F. (1958), Nobel lecture: The chemistry of insulin (PDF), Nobelprize.org, retrieved 2010-10-18. Sanger's Nobel lecture was also published in Science: Sanger 1959
- ^ The Nobel Prize in Chemistry 1958: Frederick Sanger, Nobelprize.org, retrieved 2010-10-08
- ^ Marcker & Sanger 1964
- ^ Holley, R.W.; Apgar, J.; Everett, G.A.; Madison, J.T.; Marquissee, M.; Merrill, S.H.; Penswick, J.R.; Zamir, A. (1965), "Structure of a ribonucleic acid", Science, 147 (3664): 1462–1465, doi:10.1126/science.147.3664.1462, PMID 14263761
- ^ Brownlee, Sanger & Barrell 1967; Brownlee, Sanger & Barrell 1968
- ^ Sanger et al. 1973
- ^ Sanger & Coulson 1975
- ^ a b Sanger, F. (1980), Nobel lecture: Determination of nucleotide sequences in DNA (PDF), Nobelprize.org, retrieved 2010-10-18
- ^ Sanger et al. 1977
- ^ Sanger, Nicklen & Coulson 1977
- ^ The Nobel Prize in Chemistry 1980: Paul Berg, Walter Gilbert, Frederick Sanger, Nobelprize.org, retrieved 2010-10-08
- ^ Anderson et al. 1981
- ^ Sanger et al. 1982
- ^ a b c Frederick Sanger, Wellcome Trust Sanger Institute, retrieved 2010-10-12
- ^ a b Hargittai, István (April 1999), "Interview: Frederick Sanger", The Chemical Intelligencer, 4 (2), New York: Springer-Verlag: 6–11. This interview, which took place on 16 September 1997, was republished in: Hargittai, István (2002), "Chapter 5: Frederick Sanger", Candid science II: conversations with famous biomedical scientists, London: Imperial College Press, pp. 73–83, ISBN 1-86094-288-1
- ^ a b Ahuja, Anjana (12 January 2000), "The double Nobel laureate who began the book of life", The Times, London, p. 40
- ^ Bhattachjee, Yudhijit, ed. (2007), "Newsmakers: A Life in Science", Science, 317: 879
Selected publications by Frederick Sanger
- Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 16747571, please use {{cite journal}} with
|pmid=16747571instead. - Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 16747737, please use {{cite journal}} with
|pmid=16747737instead. - Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 16747948, please use {{cite journal}} with
|pmid=16747948instead. - Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 20344639, please use {{cite journal}} with
|pmid=20344639instead. - Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 16748281, please use {{cite journal}} with
|pmid=16748281instead. - Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 16748471, please use {{cite journal}} with
|pmid=16748471instead. - Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 15396627, please use {{cite journal}} with
|pmid=15396627instead. - Sanger, F.; Tuppy, H. (1951a), "The amino-acid sequence in the phenylalanyl chain of insulin. 1. The identification of lower peptides from partial hydrolysates", Biochemical Journal, 49 (4): 463–481, PMC 1197535, PMID 14886310.
- Sanger, F.; Tuppy, H. (1951b), "The amino-acid sequence in the phenylalanyl chain of insulin. 2. The investigation of peptides from enzymic hydrolysates", Biochemical Journal, 49 (4): 481–490, PMC 1197536, PMID 14886311.
- Sanger, F.; Thompson, E.O.P. (1953a), "The amino-acid sequence in the glycyl chain of insulin. 1. The identification of lower peptides from partial hydrolysates", Biochemical Journal, 53 (3): 353–366, PMC 1198157, PMID 13032078.
- Sanger, F.; Thompson, E.O.P. (1953b), "The amino-acid sequence in the glycyl chain of insulin. 2. The investigation of peptides from enzymic hydrolysates", Biochemical Journal, 53 (3): 366–374, PMC 1198158, PMID 13032079.
- Sanger, F.; Thompson, E.O.p.; Kitai, R. (1955), "The amide groups of insulin", Biochemical Journal, 59 (3): 509–518, PMC 1216278, PMID 14363129.
- Ryle, A.P.; Sanger, F.; Smith, L.F.; Kitai, R. (1955), "The disulphide bonds of insulin", Biochemical Journal, 60 (4): 541–556, PMC 1216151, PMID 13249947.
- Brown, H.; Sanger, F.; Kitai, R. (1955), "The structure of pig and sheep insulins", Biochemical Journal, 60 (4): 556–565, PMC 1216152, PMID 13249948.
- Sanger, F. (1959), "Chemistry of Insulin: determination of the structure of insulin opens the way to greater understanding of life processes", Science, 129 (3359): 1340–1344, doi:10.1126/science.129.3359.1340, PMID 13658959.
- Milstein, C.; Sanger, F. (1961), "An amino acid sequence in the active centre of phosphoglucomutase", Biochemical Journal, 79 (3): 456–469, PMC 1205670, PMID 13771000.
- Marcker, K.; Sanger, F. (1964), "N-formyl-methionyl-S-RNA", Journal of Molecular Biology, 8 (6): 835–840, doi:10.1016/S0022-2836(64)80164-9, PMID 14187409.
- Sanger, F.; Brownlee, G.G.; Barrell, B.G. (1965), "A two-dimensional fractionation procedure for radioactive nucleotides", Journal of Molecular Biology, 13 (2): 373–398, doi:10.1016/S0022-2836(65)80104-8, PMID 5325727.
- Brownlee, G.G.; Sanger, F.; Barrell, B.G. (1967), "Nucleotide sequence of 5S-ribosomal RNA from Escherichia coli", Nature, 215 (5102): 735–736, doi:10.1038/215735a0, PMID 4862513.
- Brownlee, G.G.; Sanger, F. (1967), "Nucleotide sequences from the low molecular weight ribosomal RNA of Escherichia coli", Journal of Molecular Biology, 23 (3): 337–353, doi:10.1016/S0022-2836(67)80109-8, PMID 4291728.
- Brownlee, G.G.; Sanger, F.; Barrell, B.G. (1968), "The sequence of 5S ribosomal ribonucleic acid", Journal of Molecular Biology, 34 (3): 379–412, doi:10.1016/0022-2836(68)90168-X, PMID 4938553.
- Adams, J.M.; Jeppesen, P.G.; Sanger, F.; Barrell, B.G. (1969), "Nucleotide sequence from the coat protein cistron of R17 bacteriophage RNA", Nature, 223 (5210): 1009–1014, doi:10.1038/2231009a0, PMID 5811898.
- Barrell, B.G.; Sanger, F. (1969), "The sequence of phenylalanine tRNA from E. coli", FEBS Letters, 3 (4): 275–278, doi:10.1016/0014-5793(69)80157-2, PMID 11947028.
- Jeppesen, P.G.; Barrell, B.G.; Sanger, F.; Coulson, A.R. (1972), "Nucleotide sequences of two fragments from the coat-protein cistron of bacteriophage R17 ribonucleic acid", Biochemical Journal, 128 (5): 993–1006, PMC 1173988, PMID 4566195.
- Sanger, F.; Donelson, J.E.; Coulson, A.R.; Kössel, H.; Fischer, D. (1973), "Use of DNA Polymerase I Primed by a Synthetic Oligonucleotide to Determine a Nucleotide Sequence in Phage f1 DNA", Proceedings of the National Academy of Sciences USA, 70 (4): 1209–1213, doi:10.1073/pnas.70.4.1209, PMC 433459, PMID 4577794.
- Sanger, F.; Coulson, A.R. (1975), "A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase", Journal of Molecular Biology, 94 (3): 441–448, doi:10.1016/0022-2836(75)90213-2, PMID 1100841.
- Sanger, F.; Nicklen, S.; Coulson, A.R. (1977), "DNA sequencing with chain-terminating inhibitors", Proceedings of the National Academy of Sciences USA, 74 (12): 5463–5467, doi:10.1073/pnas.74.12.5463, PMC 431765, PMID 271968. According to the Institute for Scientific Information (ISI) database, by October 2010 this paper had been cited over 64,000 times.
- Sanger, F.; Air, G.M.; Barrell, B.G.; Brown, N.L.; Coulson, A.R.; Fiddes, C.A.; Hutchinson, C.A.; Slocombe, P.M.; Smith, M. (1977), "Nucleotide sequence of bacteriophage φX174 DNA", Nature, 265 (5596): 687–695, doi:10.1038/265687a0, PMID 870828.
- Sanger, F.; Coulson, A.R. (1978), "The use of thin acrylamide gels for DNA sequencing", FEBS Letters, 87 (1): 107–110, doi:10.1016/0014-5793(78)80145-8, PMID 631324.
- Sanger, F.; Coulson, A.R.; Barrell, B.G.; Smith, A.J.; Roe, B.A. (1980), "Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing", Journal of Molecular Biology, 143 (2): 161–178, doi:10.1016/0022-2836(80)90196-5, PMID 6260957.
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; De Bruijn, M.H.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A. (1981), "Sequence and organization of the human mitochondrial genome", Nature, 290 (5806): 457–465, doi:10.1038/290457a0, PMID 7219534.
- Anderson, S.; De Bruijn, M.H.; Coulson, A.R.; Eperon, I.C.; Sanger, F.; Young, I.G. (1982), "Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome", Journal of Molecular Biology, 156 (4): 683–717, doi:10.1016/0022-2836(82)90137-1, PMID 7120390.
- Sanger, F.; Coulson, A.R.; Hong, G.F.; Hill, D.F.; Petersen, G.B. (1982), "Nucleotide sequence of bacteriophage λ DNA", Journal of Molecular Biology, 162 (4): 729–773, doi:10.1016/0022-2836(82)90546-0, PMID 6221115.
- Sanger, F. (1988), "Sequences, sequences, and sequences", Annual Review of Biochemistry, 57: 1–28, doi:10.1146/annurev.bi.57.070188.000245, PMID 2460023.
Further reading
- Finch, John (2008), A Nobel Fellow on every floor: a history of the Medical Research Council Laboratory of Molecular Biology, Cambridge: Medical Research Council, ISBN 978-1-84046-940-0.
- Sanger, F.; Dowding, M. (1996), Selected Papers of Frederick Sanger: with commentaries, Singapore: World Scientific, ISBN 981-02-2430-3.
External links
- The Sanger Institute
- About the 1958 Nobel Prize
- About the 1980 Nobel Prize
- Fred Sanger 2001 Video Documentary by The Vega Science Trust
- National Portrait Gallery
- Frederick Sanger interviewed by Alan Macfarlane, 24th August 2007 (film)
- Interviews with Nobel Prize winning scientists: Dr Frederick Sanger, British Broadcasting Corporation, c. 1985. Interviewed by Lewis Wolpert. Duration 1 hour.
- 1918 births
- Members of the United States National Academy of Sciences
- Alumni of St John's College, Cambridge
- Members of the European Molecular Biology Organization
- Fellows of King's College, Cambridge
- Academics of the University of Cambridge
- Commanders of the Order of the British Empire
- English biochemists
- Fellows of the Royal Society
- Living people
- Members of the Order of Merit
- Members of the Order of the Companions of Honour
- Nobel laureates in Chemistry
- Nobel laureates with multiple Nobel awards
- British Nobel laureates
- People educated at Bryanston School
- Old Malvernians
- Members of the French Academy of Sciences
- British pacifists
- British conscientious objectors
- Royal Medal winners
- Recipients of the Copley Medal

