Bacteriophage MS2
| Emesvirus zinderi | |
|---|---|
| |
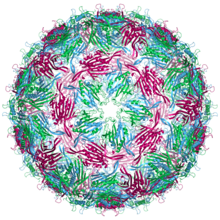
| Bacteriophage MS2 capsid structure. The three quasi-equivalent conformers are labelled blue (chain a), green (chain b) and magenta (chain c) | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Lenarviricota |
| Class: | Leviviricetes |
| Order: | Norzivirales |
| Family: | Fiersviridae |
| Genus: | Emesvirus |
| Species: | Emesvirus zinderi
|
Bacteriophage MS2 (Emesvirus zinderi), commonly called MS2, is an icosahedral, positive-sense single-stranded RNA virus that infects the bacterium Escherichia coli and other members of the Enterobacteriaceae.[1] MS2 is a member of a family of closely related bacterial viruses that includes bacteriophage f2, bacteriophage Qβ, R17, and GA.[2]
It is small and contains a maturation protein, coat protein, and genomic RNA. It also has one of the smallest known genomes, encoding four proteins.
The MS2 lifecycle involves infecting bacteria with the fertility factor, enabling the virus to attach to the pilus, though the mechanism by which the virus's RNA enters the bacterium remains unknown. Once inside, the viral RNA starts functioning as a messenger RNA to produce viral proteins. MS2 replicates its plus-strand genome by creating a minus strand RNA as a template. The virus then assembles, and the bacterial cell lyses, releasing new viruses.
The virus was isolated in 1961 and its genome was the first to be fully sequenced, in 1976, providing a crucial understanding of genetic codes. In practical applications, MS2's structural components have been used to detect RNA in living cells. The virus is also under research for potential uses in drug delivery, tumor imaging, and light harvesting. Furthermore, because of its structural similarities to noroviruses, its preferred proliferation conditions, and its lack of pathogenicity to humans, MS2 serves as a substitute in studies of norovirus disease transmission.
Virology
[edit]Genome
[edit]
| Gene | Size | Gene product | aa |
|---|---|---|---|
| mat
(MS2g1) |
1487 nt | maturation | 393 |
| cp
(MS2g2) |
510 nt | coat protein | 130 |
| lys
(MS2g3) |
295 nt | lysis protein | 75 |
| rep
(MS2g4) |
2055 nt | RNA replicase, | 545 |
The MS2 genome is one of the smallest known, consisting of 3569 nucleotides of single-stranded RNA.[3] It encodes just four proteins: the maturation protein (A-protein), the lysis (lys) protein, the coat protein (cp), and the replicase (rep) protein.[1] The gene encoding lys overlaps both the 3'-end of the upstream gene (cp) and the 5'-end of the downstream gene (rep), and was one of the first known examples of overlapping genes. The positive-stranded RNA genome serves as a messenger RNA, and is translated upon viral uncoating within the host cell. Although the four proteins are encoded by the same messenger/viral RNA, they are not all expressed at the same levels.
Structure
[edit]
An MS2 virion (viral particle) is about 27 nm in diameter, as determined by electron microscopy.[4] It consists of one copy of the maturation protein and 180 copies of the coat protein (organized as 90 dimers) arranged into an icosahedral shell with triangulation number T=3, protecting the genomic RNA inside.[5] The virion has an isoelectric point (pI) of 3.9.[6]
The structure of the coat protein is a five-stranded β-sheet with two α-helices and a hairpin. When the capsid is assembled, the helices and hairpin face the exterior of the particle, while the β-sheet faces the interior.[7]
Life cycle
[edit]MS2 infects enteric bacteria carrying the fertility (F) factor, a plasmid that allows cells to serve as DNA donors in bacterial conjugation. Genes on the F plasmid specifies the proteins of the F pilus, which includes the F-pilin protein that serves as the viral receptor. MS2 attaches to the F-pilin on the side of the pilus using its single maturation protein.
Once the viral RNA has entered the cell, it begins to function as a messenger RNA for the production of phage proteins. The gene for the most abundant protein, the coat protein, can be immediately translated. The translation start of the replicase gene is normally hidden within RNA secondary structure, but can be transiently opened as ribosomes pass through the coat protein gene. Replicase translation is also shut down once large amounts of coat protein have been made; coat protein dimers bind and stabilize the RNA "operator hairpin", blocking the replicase start. The start of the maturation protein gene is accessible in RNA being replicated but hidden within RNA secondary structure in the completed MS2 RNA; this ensures translation of only a very few copies of maturation protein per RNA. Finally, the lysis protein gene can only be initiated by ribosomes that have completed translation of the coat protein gene and "slip back" to the start of the lysis protein gene, at about a 5% frequency.[1]

Replication of the plus-strand MS2 genome requires synthesis of the complementary minus strand RNA, which can then be used as a template for synthesis of a new plus strand RNA. MS2 replication has been much less well studied than replication of the highly related bacteriophage Qβ, partly because the MS2 replicase has been difficult to isolate, but is likely to be similar.[1]
The formation of the virion is thought to be initiated by binding of maturation protein to the MS2 RNA; in fact, the complex of maturation protein and RNA is infectious. The assembly of the icosahedral shell or capsid from coat proteins can occur in the absence of RNA; however, capsid assembly is nucleated by coat protein dimer binding to the operator hairpin, and assembly occurs at much lower concentrations of coat protein when MS2 RNA is present.[1]
Bacterial lysis and release of newly formed virions occurs when sufficient lysis protein has accumulated. Lysis (L) protein forms pores in the cytoplasmic membrane, which leads to loss of membrane potential and breakdown of the cell wall.[1] The lysis protein is known to bind to DnaJ via an important P330 residue.[8] A LS dipeptide motif on the L protein is found throughout the genus Levivirus and appears to be essential to the lysis activity, although their different locations suggest that they have evolved independently.[9]
MS2 in History of Science and Use
[edit]In 1961, MS2 was isolated by Alvin John Clark and recognized as an RNA-containing phage very similar to bacteriophage f2.[10]
In 1976, the MS2 genome was the first genome to be completely sequenced.[3] This was accomplished by Walter Fiers and his team, building upon their earlier milestone in 1972 of the first gene to be completely sequenced, the MS2 coat protein.[11] These sequences were determined at the RNA level.[12] The first effort at a statistical analysis of the MS2 genome was a search for patterns in the nucleotide sequence. Several non-coding sequences were identified, however at the time of this investigation (1979), the functions of the non-coding patterns were unknown.[13]
Since 1998,[14] the MS2 operator hairpin and coat protein have found utility in the detection of RNA in living cells (see MS2 tagging). MS2 and other viral capsids are also currently under investigation as agents in drug delivery, tumor imaging, and light harvesting applications.[15]
MS2, due to its structural similarities to noroviruses, its similar optimum proliferation conditions, and non-pathogenicity to humans, has been used as substitute for noroviruses in studies of disease transmission.[16]
See also
[edit]References
[edit]- ^ a b c d e f van Duin J, Tsareva N (2006). "Single-stranded RNA phages. Chapter 15". In Calendar RL (ed.). The Bacteriophages (Second ed.). Oxford University Press. pp. 175–196. ISBN 978-0195148503.
- ^ Ni CZ, White CA, Mitchell RS, Wickersham J, Kodandapani R, Peabody DS, Ely KR (December 1996). "Crystal structure of the coat protein from the GA bacteriophage: model of the unassembled dimer". Protein Science. 5 (12): 2485–93. doi:10.1002/pro.5560051211. PMC 2143325. PMID 8976557.
- ^ a b Fiers W, Contreras R, Duerinck F, Haegeman G, Iserentant D, Merregaert J, Min Jou W, Molemans F, Raeymaekers A, Van den Berghe A, Volckaert G, Ysebaert M (April 1976). "Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene". Nature. 260 (5551): 500–7. Bibcode:1976Natur.260..500F. doi:10.1038/260500a0. PMID 1264203. S2CID 4289674.
- ^ Strauss JH, Sinsheimer RL (July 1963). "Purification and properties of bacteriophage MS2 and of its ribonucleic acid". Journal of Molecular Biology. 7: 43–54. doi:10.1016/S0022-2836(63)80017-0. PMID 13978804.
- ^ Valegård K, Liljas L, Fridborg K, Unge T (May 1990). "The three-dimensional structure of the bacterial virus MS2". Nature. 345 (6270): 36–41. Bibcode:1990Natur.345...36V. doi:10.1038/345036a0. PMID 2330049. S2CID 2803228.
- ^ Dowd SE, Pillai SD, Wang S, Corapcioglu MY (1998). "Delineating the Specific Influence of Virus Isoelectric Point and Size on Virus Adsorption and Transport Through Sandy Soils". Appl. Environ. Microbiol. 64 (2): 405–410. Bibcode:1998ApEnM..64..405D. doi:10.1128/aem.64.2.405-410.1998. PMC 106058. PMID 9464373.
- ^ Golmohammadi R, Valegård K, Fridborg K, Liljas L (December 1993). "The refined structure of bacteriophage MS2 at 2.8 A resolution". Journal of Molecular Biology. 234 (3): 620–39. doi:10.1006/jmbi.1993.1616. PMID 8254664.
- ^ Chamakura KR, Tran JS, Young R (June 2017). "MS2 Lysis of Escherichia coli Depends on Host Chaperone DnaJ". Journal of Bacteriology. 199 (12). doi:10.1128/JB.00058-17. PMC 5446614. PMID 28396351.
- ^ Chamakura KR, Edwards GB, Young R (July 2017). "Mutational analysis of the MS2 lysis protein L". Microbiology. 163 (7): 961–969. doi:10.1099/mic.0.000485. PMC 5775895. PMID 28691656.
- ^ "National Academy of Sciences: Abstracts of Papers Presented at the Autumn Meeting, 29 October, La Jolla, California, 30 October-1 November 1961, Los Angeles". Science. 134 (3488): 1425–37. November 1961. Bibcode:1961Sci...134.1425.. doi:10.1126/science.134.3488.1425. PMID 17795773.
- ^ Min Jou W, Haegeman G, Ysebaert M, Fiers W (May 1972). "Nucleotide sequence of the gene coding for the bacteriophage MS2 coat protein". Nature. 237 (5350): 82–8. Bibcode:1972Natur.237...82J. doi:10.1038/237082a0. PMID 4555447. S2CID 4153893.
- ^ Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M (February 1977). "Nucleotide sequence of bacteriophage phi X174 DNA". Nature. 265 (5596): 687–95. Bibcode:1977Natur.265..687S. doi:10.1038/265687a0. PMID 870828. S2CID 4206886.
- ^ Erickson JW, Altman GG (April 1979). "A search for patterns in the nucleotide sequence of the MS2 genome". Journal of Mathematical Biology. 7 (3): 219–30. doi:10.1007/BF00275725. S2CID 85199492.
- ^ Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM (October 1998). "Localization of ASH1 mRNA particles in living yeast". Molecular Cell. 2 (4): 437–45. doi:10.1016/S1097-2765(00)80143-4. PMID 9809065.
- ^ Glasgow J, Tullman-Ercek D (July 2014). "Production and applications of engineered viral capsids". Applied Microbiology and Biotechnology. 98 (13): 5847–58. doi:10.1007/s00253-014-5787-3. PMID 24816622. S2CID 6212583.
- ^ Fox M (8 September 2014). "Viruses spread 'like crazy' in an office, study finds". The Today Show.
External links
[edit]- Complete genome (also isolates R17, DL16, and J20)
