Chromosome
| Part of a series on |
| Genetics |
|---|
 |

- Chromatid
- Centromere
- Short arm
- Long arm
A chromosome is a package of DNA containing part or all of the genetic material of an organism. In most chromosomes, the very long thin DNA fibers are coated with nucleosome-forming packaging proteins; in eukaryotic cells, the most important of these proteins are the histones. Aided by chaperone proteins, the histones bind to and condense the DNA molecule to maintain its integrity.[1][2] These eukaryotic chromosomes display a complex three-dimensional structure that has a significant role in transcriptional regulation.[3]
Normally, chromosomes are visible under a light microscope only during the metaphase of cell division, where all chromosomes are aligned in the center of the cell in their condensed form.[4] Before this stage occurs, each chromosome is duplicated (S phase), and the two copies are joined by a centromere—resulting in either an X-shaped structure if the centromere is located equatorially, or a two-armed structure if the centromere is located distally; the joined copies are called 'sister chromatids'. During metaphase, the duplicated structure (called a 'metaphase chromosome') is highly condensed and thus easiest to distinguish and study.[5] In animal cells, chromosomes reach their highest compaction level in anaphase during chromosome segregation.[6]
Chromosomal recombination during meiosis and subsequent sexual reproduction plays a crucial role in genetic diversity. If these structures are manipulated incorrectly, through processes known as chromosomal instability and translocation, the cell may undergo mitotic catastrophe. This will usually cause the cell to initiate apoptosis, leading to its own death, but the process is occasionally hampered by cell mutations that result in the progression of cancer.
The term 'chromosome' is sometimes used in a wider sense to refer to the individualized portions of chromatin in cells, which may or may not be visible under light microscopy. In a narrower sense, 'chromosome' can be used to refer to the individualized portions of chromatin during cell division, which are visible under light microscopy due to high condensation.
Etymology
[edit]The word chromosome (/ˈkroʊməˌsoʊm, -ˌzoʊm/)[7][8] comes from the Greek words χρῶμα (chroma, "colour") and σῶμα (soma, "body"), describing the strong staining produced by particular dyes.[9] The term was coined by the German anatomist Heinrich Wilhelm Waldeyer,[10] referring to the term 'chromatin', which was introduced by Walther Flemming.
Some of the early karyological terms have become outdated.[11][12] For example, 'chromatin' (Flemming 1880) and 'chromosom' (Waldeyer 1888) both ascribe color to a non-colored state.[13]
History of discovery
[edit]Otto Bütschli was the first scientist to recognize the structures now known as chromosomes.[14]
In a series of experiments beginning in the mid-1880s, Theodor Boveri gave definitive contributions to elucidating that chromosomes are the vectors of heredity, with two notions that became known as 'chromosome continuity' and 'chromosome individuality'.[15]
Wilhelm Roux suggested that every chromosome carries a different genetic configuration, and Boveri was able to test and confirm this hypothesis. Aided by the rediscovery at the start of the 1900s of Gregor Mendel's earlier experimental work, Boveri identified the connection between the rules of inheritance and the behaviour of the chromosomes. Two generations of American cytologists were influenced by Boveri: Edmund Beecher Wilson, Nettie Stevens, Walter Sutton and Theophilus Painter (Wilson, Stevens, and Painter actually worked with him).[16]
In his famous textbook, The Cell in Development and Heredity, Wilson linked together the independent work of Boveri and Sutton (both around 1902) by naming the chromosome theory of inheritance the 'Boveri–Sutton chromosome theory' (sometimes known as the 'Sutton–Boveri chromosome theory').[17] Ernst Mayr remarks that the theory was hotly contested by some famous geneticists, including William Bateson, Wilhelm Johannsen, Richard Goldschmidt and T.H. Morgan, all of a rather dogmatic mindset. Eventually, absolute proof came from chromosome maps in Morgan's own laboratory.[18]
The number of human chromosomes was published by Painter in 1923. By inspection through a microscope, he counted 24 pairs of chromosomes, giving 48 in total. His error was copied by others, and it was not until 1956 that the true number (46) was determined by Indonesian-born cytogeneticist Joe Hin Tjio.[19]
Prokaryotes
[edit]The prokaryotes – bacteria and archaea – typically have a single circular chromosome.[20] The chromosomes of most bacteria (also called genophores), can range in size from only 130,000 base pairs in the endosymbiotic bacteria Candidatus Hodgkinia cicadicola[21] and Candidatus Tremblaya princeps,[22] to more than 14,000,000 base pairs in the soil-dwelling bacterium Sorangium cellulosum.[23]
Some bacteria have more than one chromosome. For instance, Spirochaetes such as Borrelia burgdorferi (causing Lyme disease), contain a single linear chromosome.[24] Vibrios typically carry two chromosomes of very different size. Genomes of the genus Burkholderia carry one, two, or three chromosomes.[25]
Structure in sequences
[edit]Prokaryotic chromosomes have less sequence-based structure than eukaryotes. Bacteria typically have a one-point (the origin of replication) from which replication starts, whereas some archaea contain multiple replication origins.[26] The genes in prokaryotes are often organized in operons and do not usually contain introns, unlike eukaryotes.
DNA packaging
[edit]Prokaryotes do not possess nuclei. Instead, their DNA is organized into a structure called the nucleoid.[27][28] The nucleoid is a distinct structure and occupies a defined region of the bacterial cell. This structure is, however, dynamic and is maintained and remodeled by the actions of a range of histone-like proteins, which associate with the bacterial chromosome.[29] In archaea, the DNA in chromosomes is even more organized, with the DNA packaged within structures similar to eukaryotic nucleosomes.[30][31]
Certain bacteria also contain plasmids or other extrachromosomal DNA. These are circular structures in the cytoplasm that contain cellular DNA and play a role in horizontal gene transfer.[5] In prokaryotes and viruses,[32] the DNA is often densely packed and organized; in the case of archaea, by homology to eukaryotic histones, and in the case of bacteria, by histone-like proteins.
Bacterial chromosomes tend to be tethered to the plasma membrane of the bacteria. In molecular biology application, this allows for its isolation from plasmid DNA by centrifugation of lysed bacteria and pelleting of the membranes (and the attached DNA).
Prokaryotic chromosomes and plasmids are, like eukaryotic DNA, generally supercoiled. The DNA must first be released into its relaxed state for access for transcription, regulation, and replication.
Eukaryotes
[edit]
Each eukaryotic chromosome consists of a long linear DNA molecule associated with proteins, forming a compact complex of proteins and DNA called chromatin. Chromatin contains the vast majority of the DNA in an organism, but a small amount inherited maternally can be found in the mitochondria. It is present in most cells, with a few exceptions, for example, red blood cells.
Histones are responsible for the first and most basic unit of chromosome organization, the nucleosome.
Eukaryotes (cells with nuclei such as those found in plants, fungi, and animals) possess multiple large linear chromosomes contained in the cell's nucleus. Each chromosome has one centromere, with one or two arms projecting from the centromere, although, under most circumstances, these arms are not visible as such. In addition, most eukaryotes have a small circular mitochondrial genome, and some eukaryotes may have additional small circular or linear cytoplasmic chromosomes.

In the nuclear chromosomes of eukaryotes, the uncondensed DNA exists in a semi-ordered structure, where it is wrapped around histones (structural proteins), forming a composite material called chromatin.
Interphase chromatin
[edit]The packaging of DNA into nucleosomes causes a 10 nanometer fibre which may further condense up to 30 nm fibres[33] Most of the euchromatin in interphase nuclei appears to be in the form of 30-nm fibers.[33] Chromatin structure is the more decondensed state, i.e. the 10-nm conformation allows transcription.[33]

During interphase (the period of the cell cycle where the cell is not dividing), two types of chromatin can be distinguished:
- Euchromatin, which consists of DNA that is active, e.g., being expressed as protein.
- Heterochromatin, which consists of mostly inactive DNA. It seems to serve structural purposes during the chromosomal stages. Heterochromatin can be further distinguished into two types:
- Constitutive heterochromatin, which is never expressed. It is located around the centromere and usually contains repetitive sequences.
- Facultative heterochromatin, which is sometimes expressed.
Metaphase chromatin and division
[edit]

In the early stages of mitosis or meiosis (cell division), the chromatin double helix becomes more and more condensed. They cease to function as accessible genetic material (transcription stops) and become a compact transportable form. The loops of thirty-nanometer chromatin fibers are thought to fold upon themselves further to form the compact metaphase chromosomes of mitotic cells. The DNA is thus condensed about ten-thousand-fold.[33]
The chromosome scaffold, which is made of proteins such as condensin, TOP2A and KIF4,[34] plays an important role in holding the chromatin into compact chromosomes. Loops of thirty-nanometer structure further condense with scaffold into higher order structures.[35]
This highly compact form makes the individual chromosomes visible, and they form the classic four-arm structure, a pair of sister chromatids attached to each other at the centromere. The shorter arms are called p arms (from the French petit, small) and the longer arms are called q arms (q follows p in the Latin alphabet; q-g "grande"; alternatively it is sometimes said q is short for queue meaning tail in French[36]). This is the only natural context in which individual chromosomes are visible with an optical microscope.
Mitotic metaphase chromosomes are best described by a linearly organized longitudinally compressed array of consecutive chromatin loops.[37]
During mitosis, microtubules grow from centrosomes located at opposite ends of the cell and also attach to the centromere at specialized structures called kinetochores, one of which is present on each sister chromatid. A special DNA base sequence in the region of the kinetochores provides, along with special proteins, longer-lasting attachment in this region. The microtubules then pull the chromatids apart toward the centrosomes, so that each daughter cell inherits one set of chromatids. Once the cells have divided, the chromatids are uncoiled and DNA can again be transcribed. In spite of their appearance, chromosomes are structurally highly condensed, which enables these giant DNA structures to be contained within a cell nucleus.
Human chromosomes
[edit]Chromosomes in humans can be divided into two types: autosomes (body chromosome(s)) and allosome (sex chromosome(s)). Certain genetic traits are linked to a person's sex and are passed on through the sex chromosomes. The autosomes contain the rest of the genetic hereditary information. All act in the same way during cell division. Human cells have 23 pairs of chromosomes (22 pairs of autosomes and one pair of sex chromosomes), giving a total of 46 per cell. In addition to these, human cells have many hundreds of copies of the mitochondrial genome. Sequencing of the human genome has provided a great deal of information about each of the chromosomes. Below is a table compiling statistics for the chromosomes, based on the Sanger Institute's human genome information in the Vertebrate Genome Annotation (VEGA) database.[38] Number of genes is an estimate, as it is in part based on gene predictions. Total chromosome length is an estimate as well, based on the estimated size of unsequenced heterochromatin regions.
| Chromosome | Genes[39] | Total base pairs | % of bases |
|---|---|---|---|
| 1 | 2000 | 247,199,719 | 8.0 |
| 2 | 1300 | 242,751,149 | 7.9 |
| 3 | 1000 | 199,446,827 | 6.5 |
| 4 | 1000 | 191,263,063 | 6.2 |
| 5 | 900 | 180,837,866 | 5.9 |
| 6 | 1000 | 170,896,993 | 5.5 |
| 7 | 900 | 158,821,424 | 5.2 |
| 8 | 700 | 146,274,826 | 4.7 |
| 9 | 800 | 140,442,298 | 4.6 |
| 10 | 700 | 135,374,737 | 4.4 |
| 11 | 1300 | 134,452,384 | 4.4 |
| 12 | 1100 | 132,289,534 | 4.3 |
| 13 | 300 | 114,127,980 | 3.7 |
| 14 | 800 | 106,360,585 | 3.5 |
| 15 | 600 | 100,338,915 | 3.3 |
| 16 | 800 | 88,822,254 | 2.9 |
| 17 | 1200 | 78,654,742 | 2.6 |
| 18 | 200 | 76,117,153 | 2.5 |
| 19 | 1500 | 63,806,651 | 2.1 |
| 20 | 500 | 62,435,965 | 2.0 |
| 21 | 200 | 46,944,323 | 1.5 |
| 22 | 500 | 49,528,953 | 1.6 |
| X (sex chromosome) | 800 | 154,913,754 | 5.0 |
| Y (sex chromosome) | 200[40] | 57,741,652 | 1.9 |
| Total | 21,000 | 3,079,843,747 | 100.0 |
Based on the micrographic characteristics of size, position of the centromere and sometimes the presence of a chromosomal satellite, the human chromosomes are classified into the following groups:[41][42]
| Group | Chromosomes | Features |
|---|---|---|
| A | 1–3 | Large, metacentric or submetacentric |
| B | 4–5 | Large, submetacentric |
| C | 6–12, X | Medium-sized, submetacentric |
| D | 13–15 | Medium-sized, acrocentric, with satellite |
| E | 16–18 | Small, metacentric or submetacentric |
| F | 19–20 | Very small, metacentric |
| G | 21–22, Y | Very small, acrocentric (and 21, 22 with satellite) |
Karyotype
[edit]

In general, the karyotype is the characteristic chromosome complement of a eukaryote species.[43] The preparation and study of karyotypes is part of cytogenetics.
Although the replication and transcription of DNA is highly standardized in eukaryotes, the same cannot be said for their karyotypes, which are often highly variable. There may be variation between species in chromosome number and in detailed organization. In some cases, there is significant variation within species. Often there is:
- 1. variation between the two sexes
- 2. variation between the germline and soma (between gametes and the rest of the body)
- 3. variation between members of a population, due to balanced genetic polymorphism
- 4. geographical variation between races
- 5. mosaics or otherwise abnormal individuals.
Also, variation in karyotype may occur during development from the fertilized egg.
The technique of determining the karyotype is usually called karyotyping. Cells can be locked part-way through division (in metaphase) in vitro (in a reaction vial) with colchicine. These cells are then stained, photographed, and arranged into a karyogram, with the set of chromosomes arranged, autosomes in order of length, and sex chromosomes (here X/Y) at the end.
Like many sexually reproducing species, humans have special gonosomes (sex chromosomes, in contrast to autosomes). These are XX in females and XY in males.
History and analysis techniques
[edit]Investigation into the human karyotype took many years to settle the most basic question: How many chromosomes does a normal diploid human cell contain? In 1912, Hans von Winiwarter reported 47 chromosomes in spermatogonia and 48 in oogonia, concluding an XX/XO sex determination mechanism.[44] In 1922, Painter was not certain whether the diploid number of man is 46 or 48, at first favouring 46.[45] He revised his opinion later from 46 to 48, and he correctly insisted on humans having an XX/XY system.[46]
New techniques were needed to definitively solve the problem:
- Using cells in culture
- Arresting mitosis in metaphase by a solution of colchicine
- Pretreating cells in a hypotonic solution 0.075 M KCl, which swells them and spreads the chromosomes
- Squashing the preparation on the slide forcing the chromosomes into a single plane
- Cutting up a photomicrograph and arranging the result into an indisputable karyogram.
It took until 1954 before the human diploid number was confirmed as 46.[47][48] Considering the techniques of Winiwarter and Painter, their results were quite remarkable.[49] Chimpanzees, the closest living relatives to modern humans, have 48 chromosomes as do the other great apes: in humans two chromosomes fused to form chromosome 2.
Aberrations
[edit]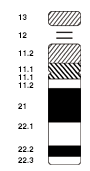
Chromosomal aberrations are disruptions in the normal chromosomal content of a cell. They can cause genetic conditions in humans, such as Down syndrome,[50] although most aberrations have little to no effect. Some chromosome abnormalities do not cause disease in carriers, such as translocations, or chromosomal inversions, although they may lead to a higher chance of bearing a child with a chromosome disorder.[citation needed] Abnormal numbers of chromosomes or chromosome sets, called aneuploidy, may be lethal or may give rise to genetic disorders.[51] Genetic counseling is offered for families that may carry a chromosome rearrangement.
The gain or loss of DNA from chromosomes can lead to a variety of genetic disorders.[52] Human examples include:
- Cri du chat, caused by the deletion of part of the short arm of chromosome 5. "Cri du chat" means "cry of the cat" in French; the condition was so-named because affected babies make high-pitched cries that sound like those of a cat. Affected individuals have wide-set eyes, a small head and jaw, moderate to severe mental health problems, and are very short.
- DiGeorge syndrome, also known as 22q11.2 deletion syndrome. Symptoms are mild learning disabilities in children, with adults having an increased risk of schizophrenia. Infections are also common in children because of problems with the immune system's T cell-mediated response due to an absence of hypoplastic thymus.[53]
- Down syndrome, the most common trisomy, usually caused by an extra copy of chromosome 21 (trisomy 21). Characteristics include decreased muscle tone, stockier build, asymmetrical skull, slanting eyes, and mild to moderate developmental disability.[54]
- Edwards syndrome, or trisomy-18, the second most common trisomy.[55] Symptoms include motor retardation, developmental disability, and numerous congenital anomalies causing serious health problems. Ninety percent of those affected die in infancy. They have characteristic clenched hands and overlapping fingers.
- Isodicentric 15, also called idic(15), partial tetrasomy 15q, or inverted duplication 15 (inv dup 15).
- Jacobsen syndrome, which is very rare. It is also called the 11q terminal deletion disorder.[56] Those affected have normal intelligence or mild developmental disability, with poor expressive language skills. Most have a bleeding disorder called Paris-Trousseau syndrome.
- Klinefelter syndrome (XXY). Men with Klinefelter syndrome are usually sterile, and tend to be taller than their peers, with longer arms and legs. Boys with the syndrome are often shy and quiet, and have a higher incidence of speech delay and dyslexia. Without testosterone treatment, some may develop gynecomastia during puberty.
- Patau Syndrome, also called D-Syndrome or trisomy-13. Symptoms are somewhat similar to those of trisomy-18, without the characteristic folded hand.
- Small supernumerary marker chromosome. This means there is an extra, abnormal chromosome. Features depend on the origin of the extra genetic material. Cat-eye syndrome and isodicentric chromosome 15 syndrome (or Idic15) are both caused by a supernumerary marker chromosome, as is Pallister–Killian syndrome.
- Triple-X syndrome (XXX). XXX girls tend to be tall and thin, and have a higher incidence of dyslexia.
- Turner syndrome (X instead of XX or XY). In Turner syndrome, female sexual characteristics are present but underdeveloped. Females with Turner syndrome often have a short stature, low hairline, abnormal eye features and bone development, and a "caved-in" appearance to the chest.
- Wolf–Hirschhorn syndrome, caused by partial deletion of the short arm of chromosome 4. It is characterized by growth retardation, delayed motor skills development, "Greek Helmet" facial features, and mild to profound mental health problems.
- XYY syndrome. XYY boys are usually taller than their siblings. Like XXY boys and XXX girls, they are more likely to have learning difficulties.
Sperm aneuploidy
[edit]Exposure of males to certain lifestyle, environmental and/or occupational hazards may increase the risk of aneuploid spermatozoa.[57] In particular, risk of aneuploidy is increased by tobacco smoking,[58][59] and occupational exposure to benzene,[60] insecticides,[61][62] and perfluorinated compounds.[63] Increased aneuploidy is often associated with increased DNA damage in spermatozoa.
Number in various organisms
[edit]In eukaryotes
[edit]The number of chromosomes in eukaryotes is highly variable. It is possible for chromosomes to fuse or break and thus evolve into novel karyotypes. Chromosomes can also be fused artificially. For example, when the 16 chromosomes of yeast were fused into one giant chromosome, it was found that the cells were still viable with only somewhat reduced growth rates.[64]
The tables below give the total number of chromosomes (including sex chromosomes) in a cell nucleus for various eukaryotes. Most are diploid, such as humans who have 22 different types of autosomes—each present as two homologous pairs—and two sex chromosomes, giving 46 chromosomes in total. Some other organisms have more than two copies of their chromosome types, for example bread wheat which is hexaploid, having six copies of seven different chromosome types for a total of 42 chromosomes.
|
|
|
Normal members of a particular eukaryotic species all have the same number of nuclear chromosomes. Other eukaryotic chromosomes, i.e., mitochondrial and plasmid-like small chromosomes, are much more variable in number, and there may be thousands of copies per cell.

Asexually reproducing species have one set of chromosomes that are the same in all body cells. However, asexual species can be either haploid or diploid.
Sexually reproducing species have somatic cells (body cells) that are diploid [2n], having two sets of chromosomes (23 pairs in humans), one set from the mother and one from the father. Gametes (reproductive cells) are haploid [n], having one set of chromosomes. Gametes are produced by meiosis of a diploid germline cell, during which the matching chromosomes of father and mother can exchange small parts of themselves (crossover) and thus create new chromosomes that are not inherited solely from either parent. When a male and a female gamete merge during fertilization, a new diploid organism is formed.
Some animal and plant species are polyploid [Xn], having more than two sets of homologous chromosomes. Important crops such as tobacco or wheat are often polyploid, compared to their ancestral species. Wheat has a haploid number of seven chromosomes, still seen in some cultivars as well as the wild progenitors. The more common types of pasta and bread are polyploid, having 28 (tetraploid) and 42 (hexaploid) chromosomes, compared to the 14 (diploid) chromosomes in wild wheat.[90]
In prokaryotes
[edit]Prokaryote species generally have one copy of each major chromosome, but most cells can easily survive with multiple copies.[91] For example, Buchnera, a symbiont of aphids has multiple copies of its chromosome, ranging from 10 to 400 copies per cell.[92] However, in some large bacteria, such as Epulopiscium fishelsoni up to 100,000 copies of the chromosome can be present.[93] Plasmids and plasmid-like small chromosomes are, as in eukaryotes, highly variable in copy number. The number of plasmids in the cell is almost entirely determined by the rate of division of the plasmid – fast division causes high copy number.
See also
[edit]Notes and references
[edit]- ^ Hammond CM, Strømme CB, Huang H, Patel DJ, Groth A (March 2017). "Histone chaperone networks shaping chromatin function". Nature Reviews. Molecular Cell Biology. 18 (3): 141–158. doi:10.1038/nrm.2016.159. PMC 5319910. PMID 28053344.
- ^ Wilson, John (2002). Molecular biology of the cell : a problems approach. New York: Garland Science. ISBN 978-0-8153-3577-1.
- ^ Bonev, Boyan; Cavalli, Giacomo (14 October 2016). "Organization and function of the 3D genome". Nature Reviews Genetics. 17 (11): 661–678. doi:10.1038/nrg.2016.112. hdl:2027.42/151884. PMID 27739532. S2CID 31259189.
- ^ Alberts B, Bray D, Hopkin K, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2014). Essential Cell Biology (Fourth ed.). New York, New York, US: Garland Science. pp. 621–626. ISBN 978-0-8153-4454-4.
- ^ a b Schleyden, M. J. (1847). Microscopical researches into the accordance in the structure and growth of animals and plants. Printed for the Sydenham Society.
- ^ Antonin W, Neumann H (June 2016). "Chromosome condensation and decondensation during mitosis" (PDF). Current Opinion in Cell Biology. 40: 15–22. doi:10.1016/j.ceb.2016.01.013. PMID 26895139.
- ^ Jones, Daniel (2003) [1917], Peter Roach; James Hartmann; Jane Setter (eds.), English Pronouncing Dictionary, Cambridge: Cambridge University Press, ISBN 978-3-12-539683-8
- ^ "Chromosome". Merriam-Webster.com Dictionary. Merriam-Webster.
- ^ Coxx, H. J. (1925). Biological Stains – A Handbook on the Nature and Uses of the Dyes Employed in the Biological Laboratory. Commission on Standardization of Biological Stains.
- ^ Waldeyer-Hartz (1888). "Über Karyokinese und ihre Beziehungen zu den Befruchtungsvorgängen". Archiv für Mikroskopische Anatomie und Entwicklungsmechanik. 32: 27.
- ^ Garbari F, Bedini G, Peruzzi L (2012). "Chromosome numbers of the Italian flora. From the Caryologia foundation to present". Caryologia – International Journal of Cytology, Cytosystematics and Cytogenetics. 65 (1): 65–66. doi:10.1080/00087114.2012.678090. S2CID 83748967.
- ^ Peruzzi L, Garbari F, Bedini G (2012). "New trends in plant cytogenetics and cytoembryology: Dedicated to the memory of Emilio Battaglia". Plant Biosystems. 146 (3): 674–675. Bibcode:2012PBios.146..674P. doi:10.1080/11263504.2012.712553. S2CID 83749502.
- ^ Battaglia, Emilio (2009). "Caryoneme alternative to chromosome and a new caryological nomenclature" (PDF). Caryologia – International Journal of Cytology, Cytosystematics. 62 (4): 1–80. Retrieved 6 November 2017.
- ^ Fokin SI (2013). "Otto Bütschli (1848–1920) Where we will genuflect?" (PDF). Protistology. 8 (1): 22–35. Archived from the original (PDF) on 21 April 2021.
- ^ Maderspacher, Florian (2008). "Theodor Boveri and the natural experiment". Current Biology. 18 (7): R279 – R286. Bibcode:2008CBio...18.R279M. doi:10.1016/j.cub.2008.02.061. PMID 18397731. S2CID 15479331.
- ^ Carlson, Elof A. (2004). Mendel's Legacy: The Origin of Classical Genetics (PDF). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. p. 88. ISBN 978-087969675-7.
- ^ Wilson, E.B. (1925). The Cell in Development and Heredity, Ed. 3. Macmillan, New York. p. 923.
- ^ Mayr, E. (1982). The growth of biological thought. Harvard. p. 749. ISBN 9780674364462
- ^ Gartler, Stanley M. (1 August 2006). "The chromosome number in humans: a brief history". Nature Reviews Genetics. 7 (8): 655–660. doi:10.1038/nrg1917. PMID 16847465. S2CID 21365693.
- ^ Thanbichler M, Shapiro L (November 2006). "Chromosome organization and segregation in bacteria". Journal of Structural Biology. 156 (2): 292–303. doi:10.1016/j.jsb.2006.05.007. PMID 16860572.
- ^ Van Leuven JT, Meister RC, Simon C, McCutcheon JP (September 2014). "Sympatric speciation in a bacterial endosymbiont results in two genomes with the functionality of one". Cell. 158 (6): 1270–1280. doi:10.1016/j.cell.2014.07.047. PMID 25175626. S2CID 11839535.
- ^ McCutcheon JP, von Dohlen CD (August 2011). "An interdependent metabolic patchwork in the nested symbiosis of mealybugs". Current Biology. 21 (16): 1366–72. Bibcode:2011CBio...21.1366M. doi:10.1016/j.cub.2011.06.051. PMC 3169327. PMID 21835622.
- ^ Han K, Li ZF, Peng R, Zhu LP, Zhou T, Wang LG, Li SG, Zhang XB, Hu W, Wu ZH, Qin N, Li YZ (2013). "Extraordinary expansion of a Sorangium cellulosum genome from an alkaline milieu". Scientific Reports. 3: 2101. Bibcode:2013NatSR...3.2101H. doi:10.1038/srep02101. PMC 3696898. PMID 23812535.
- ^ Hinnebusch J, Tilly K (December 1993). "Linear plasmids and chromosomes in bacteria". Molecular Microbiology. 10 (5): 917–22. doi:10.1111/j.1365-2958.1993.tb00963.x. PMID 7934868. S2CID 23852021.
- ^ Touchon, Marie; Rocha, Eduardo P.C. (January 2016). "Coevolution of the Organization and Structure of Prokaryotic Genomes". Cold Spring Harbor Perspectives in Biology. 8 (1): a018168. doi:10.1101/cshperspect.a018168. ISSN 1943-0264. PMC 4691797. PMID 26729648.
- ^ Kelman LM, Kelman Z (September 2004). "Multiple origins of replication in archaea". Trends in Microbiology. 12 (9): 399–401. doi:10.1016/j.tim.2004.07.001. PMID 15337158.
- ^ Thanbichler M, Wang SC, Shapiro L (October 2005). "The bacterial nucleoid: a highly organized and dynamic structure". Journal of Cellular Biochemistry. 96 (3): 506–21. doi:10.1002/jcb.20519. PMID 15988757. S2CID 25355087.
- ^ Le TB, Imakaev MV, Mirny LA, Laub MT (November 2013). "High-resolution mapping of the spatial organization of a bacterial chromosome". Science. 342 (6159): 731–4. Bibcode:2013Sci...342..731L. doi:10.1126/science.1242059. PMC 3927313. PMID 24158908.
- ^ Sandman K, Pereira SL, Reeve JN (December 1998). "Diversity of prokaryotic chromosomal proteins and the origin of the nucleosome". Cellular and Molecular Life Sciences. 54 (12): 1350–64. doi:10.1007/s000180050259. PMC 11147202. PMID 9893710. S2CID 21101836.
- ^ Sandman K, Reeve JN (March 2000). "Structure and functional relationships of archaeal and eukaryal histones and nucleosomes". Archives of Microbiology. 173 (3): 165–9. Bibcode:2000ArMic.173..165S. doi:10.1007/s002039900122. PMID 10763747. S2CID 28946064.
- ^ Pereira SL, Grayling RA, Lurz R, Reeve JN (November 1997). "Archaeal nucleosomes". Proceedings of the National Academy of Sciences of the United States of America. 94 (23): 12633–7. Bibcode:1997PNAS...9412633P. doi:10.1073/pnas.94.23.12633. PMC 25063. PMID 9356501.
- ^ Johnson JE, Chiu W (April 2000). "Structures of virus and virus-like particles". Current Opinion in Structural Biology. 10 (2): 229–35. doi:10.1016/S0959-440X(00)00073-7. PMID 10753814.
- ^ a b c d Cooper, G.M. (2019). The Cell (8 ed.). Oxford University Press. ISBN 978-1605357072.
- ^ Poonperm, Rawin; Takata, Hideaki; Hamano, Tohru; Matsuda, Atsushi; Uchiyama, Susumu; Hiraoka, Yasushi; Fukui, Kiichi (1 July 2015). "Chromosome Scaffold is a Double-Stranded Assembly of Scaffold Proteins". Scientific Reports. 5 (1): 11916. Bibcode:2015NatSR...511916P. doi:10.1038/srep11916. PMC 4487240. PMID 26132639.
- ^ Lodish, U.H.; Lodish, H.; Berk, A.; Kaiser, C.A.; Kaiser, C.; Kaiser, U.C.A.; Krieger, M.; Scott, M.P.; Bretscher, A.; Ploegh, H.; others (2008). Molecular Cell Biology. W. H. Freeman. ISBN 978-0-7167-7601-7.
- ^ "Chromosome Mapping: Idiograms" Nature Education – 13 August 2013
- ^ Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J (November 2013). "Organization of the mitotic chromosome". Science. 342 (6161): 948–53. Bibcode:2013Sci...342..948N. doi:10.1126/science.1236083. PMC 4040465. PMID 24200812.
- ^ Vega.sanger.ad.uk, all data in this table was derived from this database, 11 November 2008.
- ^ "Ensembl genome browser 71: Homo sapiens – Chromosome summary – Chromosome 1: 1–1,000,000". apr2013.archive.ensembl.org. Retrieved 11 April 2016.
- ^ "Chromosome Map". Genes and Disease. Bethesda, Maryland: National Center for Biotechnology Information. 1998.
- ^ The colors of each row match those of the karyogram (see Karyotype section)
- ^ Erwinsyah, R.; Riandi; Nurjhani, M. (2017). "Relevance of human chromosome analysis activities against mutation concept in genetics course. IOP Conference Series". Materials Science and Engineering. doi:10.1088/1757-899x/180/1/012285. S2CID 90739754.
- ^ White, M. J. D. (1973). The chromosomes (6th ed.). London: Chapman and Hall, distributed by Halsted Press, New York. p. 28. ISBN 978-0-412-11930-9.
- ^ von Winiwarter H (1912). "Études sur la spermatogenèse humaine". Archives de Biologie. 27 (93): 147–9.
- ^ Painter TS (1922). "The spermatogenesis of man". Anat. Res. 23: 129.
- ^ Painter, Theophilus S. (April 1923). "Studies in mammalian spermatogenesis. II. The spermatogenesis of man". Journal of Experimental Zoology. 37 (3): 291–336. Bibcode:1923JEZ....37..291P. doi:10.1002/jez.1400370303.
- ^ Tjio JH, Levan A (1956). "The chromosome number of man". Hereditas. 42 (1–2): 723–4. doi:10.1111/j.1601-5223.1956.tb03010.x. hdl:10261/15776. PMID 345813.
- ^ Ford CE, Hamerton JL (November 1956). "The chromosomes of man". Nature. 178 (4541): 1020–3. Bibcode:1956Natur.178.1020F. doi:10.1038/1781020a0. PMID 13378517. S2CID 4155320.
- ^ Hsu T.C. (1979) Human and mammalian cytogenetics: a historical perspective. Springer-Verlag, N.Y. ISBN 9780387903644 p. 10: "It's amazing that he [Painter] even came close!"
- ^ "Chromosomal Abnormalities", Understanding Genetics: A New York, Mid-Atlantic Guide for Patients and Health Professionals, Genetic Alliance, 8 July 2009, retrieved 27 September 2023
- ^ Santaguida S, Amon A (August 2015). "Short- and long-term effects of chromosome mis-segregation and aneuploidy" (PDF). Nature Reviews. Molecular Cell Biology. 16 (8): 473–85. doi:10.1038/nrm4025. hdl:1721.1/117201. PMID 26204159. S2CID 205495880.
- ^ "Genetic Disorders". medlineplus.gov. Retrieved 27 April 2022.
- ^ "DiGeorge Syndrome". www.ncbi.nlm.nih.gov. Retrieved 8 August 2023.
- ^ Miller KR (2000). "Chapter 9-3". Biology (5th ed.). Upper Saddle River, New Jersey: Prentice Hall. pp. 194–5. ISBN 978-0-13-436265-6.
- ^ "What is Trisomy 18?". Trisomy 18 Foundation. Archived from the original on 30 January 2017. Retrieved 4 February 2017.
- ^ "Terminal deletion". European Chromosome 11 Network. Retrieved 20 February 2023.
- ^ Templado C, Uroz L, Estop A (October 2013). "New insights on the origin and relevance of aneuploidy in human spermatozoa". Molecular Human Reproduction. 19 (10): 634–43. doi:10.1093/molehr/gat039. PMID 23720770.
- ^ Shi Q, Ko E, Barclay L, Hoang T, Rademaker A, Martin R (August 2001). "Cigarette smoking and aneuploidy in human sperm". Molecular Reproduction and Development. 59 (4): 417–21. doi:10.1002/mrd.1048. PMID 11468778. S2CID 35230655.
- ^ Rubes J, Lowe X, Moore D, Perreault S, Slott V, Evenson D, Selevan SG, Wyrobek AJ (October 1998). "Smoking cigarettes is associated with increased sperm disomy in teenage men". Fertility and Sterility. 70 (4): 715–23. doi:10.1016/S0015-0282(98)00261-1. PMID 9797104.
- ^ Xing C, Marchetti F, Li G, Weldon RH, Kurtovich E, Young S, Schmid TE, Zhang L, Rappaport S, Waidyanatha S, Wyrobek AJ, Eskenazi B (June 2010). "Benzene exposure near the U.S. permissible limit is associated with sperm aneuploidy". Environmental Health Perspectives. 118 (6): 833–9. Bibcode:2010EnvHP.118..833X. doi:10.1289/ehp.0901531. PMC 2898861. PMID 20418200.
- ^ Xia Y, Bian Q, Xu L, Cheng S, Song L, Liu J, Wu W, Wang S, Wang X (October 2004). "Genotoxic effects on human spermatozoa among pesticide factory workers exposed to fenvalerate". Toxicology. 203 (1–3): 49–60. Bibcode:2004Toxgy.203...49X. doi:10.1016/j.tox.2004.05.018. PMID 15363581. S2CID 36073841.
- ^ Xia Y, Cheng S, Bian Q, Xu L, Collins MD, Chang HC, Song L, Liu J, Wang S, Wang X (May 2005). "Genotoxic effects on spermatozoa of carbaryl-exposed workers". Toxicological Sciences. 85 (1): 615–23. doi:10.1093/toxsci/kfi066. PMID 15615886.
- ^ Governini L, Guerranti C, De Leo V, Boschi L, Luddi A, Gori M, Orvieto R, Piomboni P (November 2015). "Chromosomal aneuploidies and DNA fragmentation of human spermatozoa from patients exposed to perfluorinated compounds". Andrologia. 47 (9): 1012–9. doi:10.1111/and.12371. hdl:11365/982323. PMID 25382683. S2CID 13484513.
- ^ Shao, Yangyang; Lu, Ning; Wu, Zhenfang; Cai, Chen; Wang, Shanshan; Zhang, Ling-Li; Zhou, Fan; Xiao, Shijun; Liu, Lin; Zeng, Xiaofei; Zheng, Huajun (August 2018). "Creating a functional single-chromosome yeast". Nature. 560 (7718): 331–335. Bibcode:2018Natur.560..331S. doi:10.1038/s41586-018-0382-x. ISSN 1476-4687. PMID 30069045. S2CID 51894920.
- ^ Armstrong SJ, Jones GH (January 2003). "Meiotic cytology and chromosome behaviour in wild-type Arabidopsis thaliana". Journal of Experimental Botany. 54 (380): 1–10. doi:10.1093/jxb/54.380.1. PMID 12456750.
- ^ Gill BS, Kimber G (April 1974). "The Giemsa C-banded karyotype of rye". Proceedings of the National Academy of Sciences of the United States of America. 71 (4): 1247–9. Bibcode:1974PNAS...71.1247G. doi:10.1073/pnas.71.4.1247. PMC 388202. PMID 4133848.
- ^ a b c Dubcovsky J, Luo MC, Zhong GY, Bransteitter R, Desai A, Kilian A, Kleinhofs A, Dvorák J (June 1996). "Genetic map of diploid wheat, Triticum monococcum L., and its comparison with maps of Hordeum vulgare L". Genetics. 143 (2): 983–99. doi:10.1093/genetics/143.2.983. PMC 1207354. PMID 8725244.
- ^ Kato A, Lamb JC, Birchler JA (September 2004). "Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize". Proceedings of the National Academy of Sciences of the United States of America. 101 (37): 13554–9. Bibcode:2004PNAS..10113554K. doi:10.1073/pnas.0403659101. PMC 518793. PMID 15342909.
- ^ Kenton A, Parokonny AS, Gleba YY, Bennett MD (August 1993). "Characterization of the Nicotiana tabacum L. genome by molecular cytogenetics". Molecular & General Genetics. 240 (2): 159–69. doi:10.1007/BF00277053. PMID 8355650. S2CID 6953185.
- ^ Leitch IJ, Soltis DE, Soltis PS, Bennett MD (January 2005). "Evolution of DNA amounts across land plants (embryophyta)". Annals of Botany. 95 (1): 207–17. doi:10.1093/aob/mci014. PMC 4246719. PMID 15596468.
- ^ Ambarish, C.N.; Sridhar, K.R. (2014). "Cytological and karyological observations on two endemic giant pill-millipedes Arthrosphaera (Pocock 1895) (Diplopoda: Sphaerotheriida) of the Western Ghats of India". Caryologia. 67 (1): 49–56. doi:10.1080/00087114.2014.891700. S2CID 219554731.
- ^ Vitturi R, Colomba MS, Pirrone AM, Mandrioli M (2002). "rDNA (18S–28S and 5S) colocalization and linkage between ribosomal genes and (TTAGGG)(n) telomeric sequence in the earthworm, Octodrilus complanatus (Annelida: Oligochaeta: Lumbricidae), revealed by single- and double-color FISH". The Journal of Heredity. 93 (4): 279–82. doi:10.1093/jhered/93.4.279. PMID 12407215.
- ^ Nie W, Wang J, O'Brien PC, Fu B, Ying T, Ferguson-Smith MA, Yang F (2002). "The genome phylogeny of domestic cat, red panda and five mustelid species revealed by comparative chromosome painting and G-banding". Chromosome Research. 10 (3): 209–22. doi:10.1023/A:1015292005631. PMID 12067210. S2CID 9660694.
- ^ a b Romanenko SA, Perelman PL, Serdukova NA, Trifonov VA, Biltueva LS, Wang J, Li T, Nie W, O'Brien PC, Volobouev VT, Stanyon R, Ferguson-Smith MA, Yang F, Graphodatsky AS (December 2006). "Reciprocal chromosome painting between three laboratory rodent species". Mammalian Genome. 17 (12): 1183–92. doi:10.1007/s00335-006-0081-z. PMID 17143584. S2CID 41546146.
- ^ a b Painter TS (March 1928). "A Comparison of the Chromosomes of the Rat and Mouse with Reference to the Question of Chromosome Homology in Mammals". Genetics. 13 (2): 180–9. doi:10.1093/genetics/13.2.180. PMC 1200977. PMID 17246549.
- ^ Hayes H, Rogel-Gaillard C, Zijlstra C, De Haan NA, Urien C, Bourgeaux N, Bertaud M, Bosma AA (2002). "Establishment of an R-banded rabbit karyotype nomenclature by FISH localization of 23 chromosome-specific genes on both G- and R-banded chromosomes". Cytogenetic and Genome Research. 98 (2–3): 199–205. doi:10.1159/000069807. PMID 12698004. S2CID 29849096.
- ^ "The Genetics of the Popular Aquarium Pet – Guppy Fish". Archived from the original on 31 May 2023. Retrieved 6 December 2009.
- ^ a b De Grouchy J (August 1987). "Chromosome phylogenies of man, great apes, and Old World monkeys". Genetica. 73 (1–2): 37–52. doi:10.1007/bf00057436. PMID 3333352. S2CID 1098866.
- ^ Robinson TJ, Yang F, Harrison WR (2002). "Chromosome painting refines the history of genome evolution in hares and rabbits (order Lagomorpha)". Cytogenetic and Genome Research. 96 (1–4): 223–7. doi:10.1159/000063034. PMID 12438803. S2CID 19327437.
- ^ Chapman JA, Flux JE (1990), "section 4.W4", Rabbits, Hares and Pikas. Status Survey and Conservation Action Plan, pp. 61–94, ISBN 9782831700199
- ^ Vitturi R, Libertini A, Sineo L, Sparacio I, Lannino A, Gregorini A, Colomba M (2005). "Cytogenetics of the land snails Cantareus aspersus and C. mazzullii (Mollusca: Gastropoda: Pulmonata)". Micron. 36 (4): 351–7. doi:10.1016/j.micron.2004.12.010. PMID 15857774.
- ^ Yasukochi Y, Ashakumary LA, Baba K, Yoshido A, Sahara K (July 2006). "A second-generation integrated map of the silkworm reveals synteny and conserved gene order between lepidopteran insects". Genetics. 173 (3): 1319–28. doi:10.1534/genetics.106.055541. PMC 1526672. PMID 16547103.
- ^ Houck ML, Kumamoto AT, Gallagher DS, Benirschke K (2001). "Comparative cytogenetics of the African elephant (Loxodonta africana) and Asiatic elephant (Elephas maximus)". Cytogenetics and Cell Genetics. 93 (3–4): 249–52. doi:10.1159/000056992. PMID 11528120. S2CID 23529399.
- ^ Semba U, Umeda Y, Shibuya Y, Okabe H, Tanase S, Yamamoto T (October 2004). "Primary structures of guinea pig high- and low-molecular-weight kininogens". International Immunopharmacology. 4 (10–11): 1391–400. doi:10.1016/j.intimp.2004.06.003. PMID 15313436.
- ^ Wayne RK, Ostrander EA (March 1999). "Origin, genetic diversity, and genome structure of the domestic dog". BioEssays. 21 (3): 247–57. doi:10.1002/(SICI)1521-1878(199903)21:3<247::AID-BIES9>3.0.CO;2-Z. PMID 10333734. S2CID 5547543.
- ^ Ciudad J, Cid E, Velasco A, Lara JM, Aijón J, Orfao A (May 2002). "Flow cytometry measurement of the DNA contents of G0/G1 diploid cells from three different teleost fish species". Cytometry. 48 (1): 20–5. doi:10.1002/cyto.10100. PMID 12116377.
- ^ Burt DW (2002). "Origin and evolution of avian microchromosomes". Cytogenetic and Genome Research. 96 (1–4): 97–112. doi:10.1159/000063018. PMID 12438785. S2CID 26017998.
- ^ Itoh M, Ikeuchi T, Shimba H, Mori M, Sasaki M, Makino S (1969). "A Comparative Karyotype Study in Fourteen Species of Birds". The Japanese Journal of Genetics. 44 (3): 163–170. doi:10.1266/jjg.44.163.
- ^ Smith J, Burt DW (August 1998). "Parameters of the chicken genome (Gallus gallus)". Animal Genetics. 29 (4): 290–4. doi:10.1046/j.1365-2052.1998.00334.x. PMID 9745667.
- ^ Sakamura, Tetsu (1918). "Kurze Mitteilung über die Chromosomenzahlen und die Verwandtschaftsverhältnisse der Triticum-Arten". Shokubutsugaku Zasshi. 32 (379): 150–3. doi:10.15281/jplantres1887.32.379_150.
- ^ Charlebois R.L. (ed) 1999. Organization of the prokaryote genome. ASM Press, Washington DC.
- ^ Komaki K, Ishikawa H (March 2000). "Genomic copy number of intracellular bacterial symbionts of aphids varies in response to developmental stage and morph of their host". Insect Biochemistry and Molecular Biology. 30 (3): 253–8. Bibcode:2000IBMB...30..253K. doi:10.1016/S0965-1748(99)00125-3. PMID 10732993.
- ^ Mendell JE, Clements KD, Choat JH, Angert ER (May 2008). "Extreme polyploidy in a large bacterium". Proceedings of the National Academy of Sciences of the United States of America. 105 (18): 6730–4. Bibcode:2008PNAS..105.6730M. doi:10.1073/pnas.0707522105. PMC 2373351. PMID 18445653.
External links
[edit]- An Introduction to DNA and Chromosomes from HOPES: Huntington's Outreach Project for Education at Stanford
- Chromosome Abnormalities at AtlasGeneticsOncology
- On-line exhibition on chromosomes and genome (SIB)
- What Can Our Chromosomes Tell Us?, from the University of Utah's Genetic Science Learning Center
- Try making a karyotype yourself, from the University of Utah's Genetic Science Learning Center
- Kimballs Chromosome pages
- Chromosome News from Genome News Network
- Eurochromnet, European network for Rare Chromosome Disorders on the Internet
- Ensembl.org, Ensembl project, presenting chromosomes, their genes and syntenic loci graphically via the web
- Genographic Project Archived 12 July 2007 at the Wayback Machine
- Home reference on Chromosomes from the U.S. National Library of Medicine
- Visualisation of human chromosomes and comparison to other species
- Unique – The Rare Chromosome Disorder Support Group Support for people with rare chromosome disorders



