Pyrrolnitrin
| |
| Names | |
|---|---|
| Preferred IUPAC name
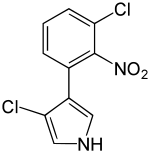
3-Chloro-4-(3-chloro-2-nitrophenyl)-1H-pyrrole | |
| Identifiers | |
3D model (JSmol)
|
|
| ChEBI | |
| ChemSpider | |
| ECHA InfoCard | 100.012.557 |
| EC Number |
|
| KEGG | |
| MeSH | D011764 |
PubChem CID
|
|
| UNII | |
CompTox Dashboard (EPA)
|
|
| |
| |
| Properties | |
| C10H6Cl2N2O2 | |
| Molar mass | 257.07284 |
| Pharmacology | |
| D01AA07 (WHO) | |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
Pyrrolnitrin (PRN[1]) is a phenylpyrrole chemical used an antifungal antibiotic.[2] Pseudomonas pyrrocinia and other Pseudomonas species produce pyrrolnitrin from tryptophan as secondary metabolite.[3][4] It is believed that the antifungal properties come from inhibition of electron transport system.[5]
The synthetic fungicides fenpiclonil and fludioxonil are chemically related to pyrrolnitrin.[6][7]
Biosynthesis
[edit]In Pseudomonas fluorescens, biosynthesis of pyrrolnitrin requires four genes, named prnABCD, arranged into a single operon. The products of these genes are similar in size and catalyze four subsequent reactions:[1][5]
- prnA – chlorination of L-tryptophan to 7-chloro-L-tryptophan (7-CLT), requiring NAD for its activity
- prnB – ring rearrangement and decarboxylation of 7-chloro-L-tryptophan to form monodechloroaminopyrrolnitrin (MAD)
- prnC – chlorination of monodechloroaminopyrrolnitrin to form aminopyrrolnitrin (APRN), requiring NAD for its activity
- prnD – oxidation of amino group to form nitro group of pyrrolnitrin

Except for prnA, these enzymes are unable to act on D-tryptophan.[1][5]
Neither of the chlorinating enzymes, prnA nor prnC, show homology to known haloperoxidases nor to one another.[1]
An alternative pathway was also suggested, where L-tryptophan is first turned into aminophenylpyrrole (APP) and then by subsequent steps to aminopyrrolnitrin and pyrrolnitrin. While these steps have not been described in more detail, prnB is able to produce APP, presumably from tryptophan as starting material.[1] APP seems to be an unwanted side product. The gene coding for prnB also starts with the unusual GTG start codon, further lowering the amount of prnB expressed and thus lowering the amount of present APP.
References
[edit]- ^ a b c d e Kirner, Sabine; Hammer, Philip E.; Hill, D. Steven; Altmann, Annett; Fischer, Ilona; Weislo, Laura J.; Lanahan, Mike; van Pée, Karl-Heinz; Ligon, James M. (April 1998). "Functions Encoded by Pyrrolnitrin Biosynthetic Genes from Pseudomonas fluorescens". Journal of Bacteriology. 180 (7): 1939–1943. doi:10.1128/JB.180.7.1939-1943.1998. ISSN 0021-9193. PMC 107110. PMID 9537395.
- ^ Gordee, R. S.; Matthews, T. R. (1969). "Systemic antifungal activity of pyrrolnitrin". Applied Microbiology. 17 (5): 690–694. doi:10.1128/AEM.17.5.690-694.1969. PMC 377781. PMID 5785951.
- ^ Zhu, X.; Van Pee, K. -H.; Naismith, J. H. (2010). "The Ternary Complex of PrnB (the Second Enzyme in the Pyrrolnitrin Biosynthesis Pathway), Tryptophan, and Cyanide Yields New Mechanistic Insights into the Indolamine Dioxygenase Superfamily". Journal of Biological Chemistry. 285 (27): 21126–21133. doi:10.1074/jbc.M110.120485. PMC 2898318. PMID 20421301.
- ^ Park, J. Y.; Oh, S. A.; Anderson, A. J.; Neiswender, J.; Kim, J. -C.; Kim, Y. C. (2011). "Production of the antifungal compounds phenazine and pyrrolnitrin from Pseudomonas chlororaphis O6 is differentially regulated by glucose". Letters in Applied Microbiology. 52 (5): 532–537. doi:10.1111/j.1472-765X.2011.03036.x. PMID 21362001.
- ^ a b c De Laurentis, Walter; Khim, Leang; Anderson, J. L. Ross; Adam, Ariane; Phillips, Robert S.; Chapman, Stephen K.; van Pee, Karl-Heinz; Naismith, James H. (2007-10-01). "The Second Enzyme in Pyrrolnitrin Biosynthetic Pathway Is Related to the Heme-Dependent Dioxygenase Superfamily". Biochemistry. 46 (43): 12393–12404. doi:10.1021/bi7012189. ISSN 0006-2960. PMC 3326534. PMID 17924666.
- ^ Pillonel, Ch; Knauf-beiter, G.; Steinemann, A. (2003). "Fungicides, Phenylpyrroles". Encyclopedia of Agrochemicals. doi:10.1002/047126363X.agr106. ISBN 047126363X.
- ^ Jespers, A.B.K.; Davidse, L.C.; Dewaard, M.A. (1993). "Biochemical Effects of the Phenylpyrrole Fungicide Fenpiclonil in Fusarium sulphureum (Schlecht)". Pesticide Biochemistry and Physiology. 45 (2): 116–129. doi:10.1006/pest.1993.1014. ISSN 0048-3575.
